Figure 6. Properties of KSHV miRNA targets.
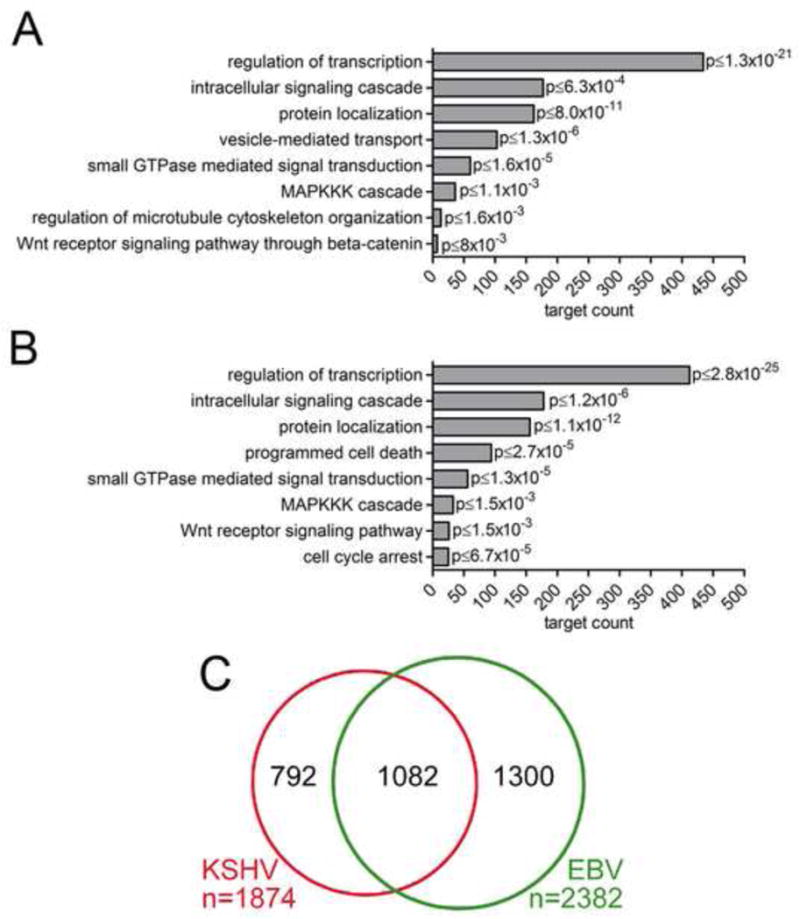
(A) Selected pathways with significant enrichments of KSHV miRNA targets. p values shown were calculated against the background of the human genome, but remained significant (p<0.05) when calculated against mRNAs expressed in both PEL cell lines or transcripts with PAR-CLIP clusters. (B) Selected pathways with significant enrichments of EBV miRNA targets. p values were calculated against human genome, all pathways shown remained significant when calculated against expressed mRNAs or transcripts with PAR-CLIP clusters in BC-1 cells. (C) Overlap of KSHV and EBV 3′UTR target mRNAs with T to C conversions at minimally 2 locations identified in BC-1. See also Tables S8, 12.
