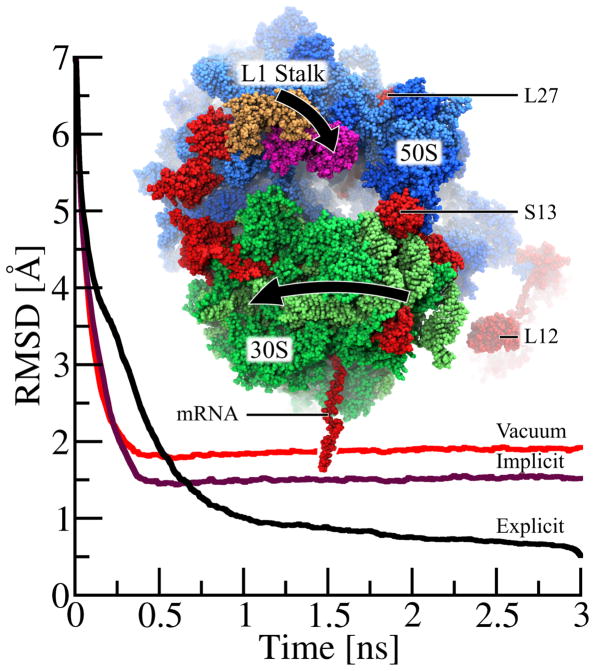
Figure 4.
Molecular dynamics flexible fitting (MDFF) of ribosome with NAMD’s GBIS method. While matching the 250,000-atom classical ribosome structure into the EM map of a ratcheted ribosome, the 30S subunit (green) rotates relative to the 50S subunit (blue) and the L1 stalk moves 30 Å from its classical (tan) to its ratcheted (magenta) position. Highlighted (red) are regions where the implicit solvent structure agrees with the explicit solvent structure much more closely than does the in vacuo structure. The root-mean-squared deviation (RMSDsol,exp(t)) of the ribosome, defined in eq. 14, with the final fitted explicit solvent structure as reference, is plotted over time for explicit solvent (RMSDexp,exp(t) in black), implicit solvent (RMSDimp,exp(t) in purple) and in vacuo (RMSDvac,exp(t) in red) MDFF. While the explicit solvent MDFF calculation requires ~1.5–2 ns to converge to its final structure, both implicit solvent and vacuum MDFF calculation require only 0.5 ns to converge. As seen by the lower RMSD values for t > 0.5 ns, the structure derived from the implicit solvent fitting agrees more closely with the final explicit solvent structure than does the in vacuo structure. While this plot illustrates only the overall improvement of the implicit solvent structure over the in vacuo structure, the text discusses key examples of ribosomal proteins (L27, S13 and L12) whose structural quality is significantly improved by the use of implicit solvent.