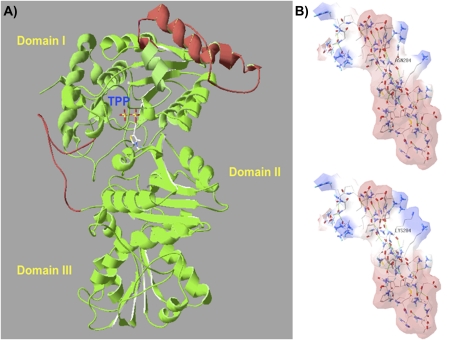
Fig. 3.
Three-dimensional VvDXS protein modelling and the putative electrostatic change conferred by the N–K substitution. DXS protein is a dimer and the structure of its monomers can be divided into three domains (I, II, and III). The active site is located between domain I and II, and the thiamine pyrophosphate coenzyme (TPP) is placed on the interface and interacts on domain II as described by Xiang et al. (2007). (A) The complete three-dimensional model of VvDXS obtained by the I-TASSER server. Two segments were reconstructed by ab initio modelling using the DXS crystallized structure (2O1×A) of Deinococcus radiodurans as template. Red parts represent linkers modelled, and mutation K284N falls within an α-helix of 46 amino acids in a linker between β-sheet 4 and 5 of domain I. (B) Electrostatic potential surface of the two VvDXS allelic forms calculated by SPDBV 4.01. Red and blue surfaces indicate the negative and positive potential, respectively.