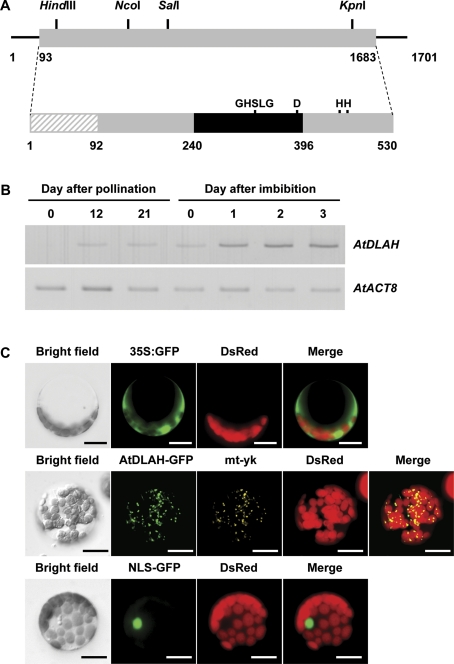
Fig. 1.
Structure, expression, and subcellular localization of Arabidopsis AtDLAH. (A) Schematic representation of AtDLAH (At1g30370) cDNA and its deduced protein. Solid lines depict the 5'- and 3'-untranslated regions. The coding region (grey box) with restriction enzyme sites, the lipase 3 domain (black box), and the putative N-terminal transit peptide (hatched box) are represented. The lipase consensus sequence (GHSLG) and the catalytic triad (serine, aspartate, and two candidate histidine residues) are indicated. (B) AtDLAH expression in developing and early germinating seeds was analysed by RT-PCR. AtACT8 was used as a loading control. (C) Subcellular localization of AtDLAH in Arabidopsis protoplasts. The 35S:AtDLAH-GFP fusion gene was introduced into protoplasts using a PEG-mediated method. The 35S:GFP and 35S:NLS-GFP constructs were used as controls for cytosolic and nuclear proteins, respectively. The mt-yk plant was used as a mitochondria-localized marker. Scale bars=10 μm. (This figure is available in colour at JXB online.)