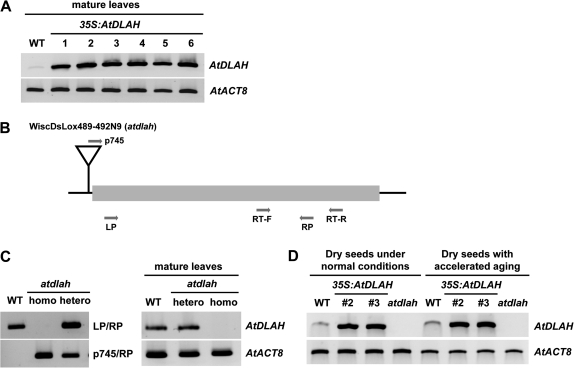
Fig. 3.
Molecular characterization of wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants. (A) RT-PCR analysis of wild-type and six independent AtDLAH-overexpressing T4 transgenic plants (lines #1, #2, #3, #4, #5, and #6) in leaves. The AtACT8 gene was used as a loading control. (B) Schematic representation of the atdlah mutant. The T-DNA insertion is shown as an inverted triangle. The shaded bar indicates the coding region, and gene-specific (LP and RP) and T-DNA-specific (p745) primers used for genotyping and RT-PCR are indicated with arrows. (C) PCR analysis of the atdlah mutant in leaves. Genomic PCR analysis of the atdlah loss-of-function mutant using gene-specific and T-DNA-specific primers (left panel). RT-PCR analysis of the atdlah mutant (right panel). The AtACT8 gene was used as a loading control. DNA sequences of primers used in this study are shown in Supplementary Table S1 at JXB online. (D) RT-PCR analysis of wild-type, 35S:AtDLAH (transgenic lines #2 and #3), and atdlah mutant seeds without or with accelerated-ageing treatment. The AtACT8 gene was used as a loading control.