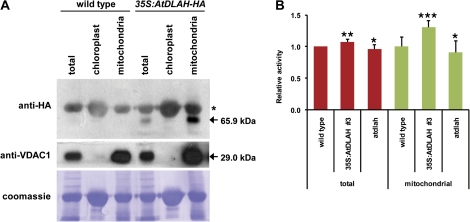
Fig. 4.
Mitochondrial localization of AtDLAH-HA and mitochondrial lipase activities in wild-type, AtDLAH-overexpressing T4 transgenic, and atdlah mutant plants. (A) Cellular fractionation analysis of the AtDLAH-HA fusion protein. Extracts of total, chloroplast, and mitochondrial fractions were prepared from wild-type and 35S:AtDLAH-HA T4 transgenic plants. Total proteins from each fraction were analysed with anti-HA and anti-VDAC1 antibodies (control for mitochondrial proteins). Loaded proteins were visualized by Coomassie staining. Arrows indicate AtDLAH-HA (65.9 kDa) and VDAC1 (29.0 kDa) proteins. An asterisk indicates non-specific binding to a 70 kDa protein by the anti-HA antibody. (B) Mitochondrial lipase enzyme assays. Total and mitochondrial proteins were prepared from wild-type, AtDLAH-overexpressing transgenic (line #3), and atdlah mutant plants and incubated with 1-palmitoyl-2-[14C]palmitoyl-PC as the substrate at 30 °C for 30 min. Lipase activities were determined by quantifying the release of 14C-labelled lyso-PC as described in Fig. 2A. Results are expressed as the means ±SD from three independent experiments. The data were analysed by Student's t-test. The statistical significance was determined at ***P <0.01, **P <0.05, and *P <0.1, respectively. (This figure is available in colour at JXB online.)