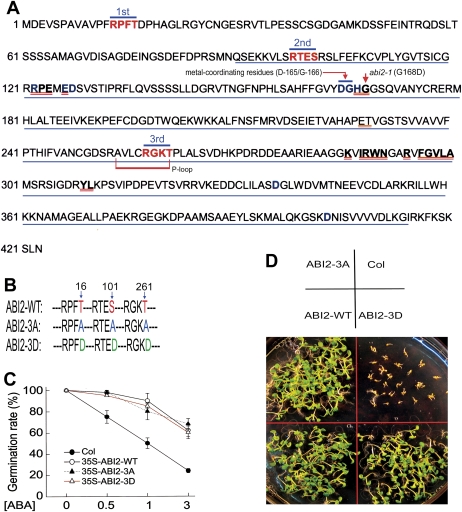
Fig. 1.
ABI2 is unlikely to function as a phosphorylated form catalysed by CDPK-mediated phosphorylation in ABA signalling. (A) Diagram of some potentially functional domains in the ABI2 molecule. Red letters indicate the three putative CDPK phosphorylation sites, and 1st, 2nd, and 3rd indicate, respectively, the first, second, and third site. Blue letters in bold indicate amino acid residues important for PP2C phosphatase activity. The sequence underlined in blue indicates the PP2C catalytic domain. Letters double-underlined in red denote the amino acid residues essential for interaction between PYL1 and ABI2 as described previously (Miyazono et al., 2009). The ABI2 metal-coordinating residues (D-165/G-166), a potential ATP/GTP-binding site motif or phosphate-binding loop (P-loop: AVLCRGKT), and the point mutation of abi2-1 (G168D) are also indicated. (B) A diagram showing the ABI2-3A (substitution mutation T16A plus S101A plus T261A) and ABI2-3D (substitution mutation T16D plus S101D plus T261D) mutations compared with wild-type ABI2 (ABI2-WT). The numbers of the serine (S)/threonine (T) residues, T16, S101, and T261, which may putatively be phosphorylated by CDPK, are indicated (blue arrows). (C) ABI2-3A and ABI2-3D transgenic lines (with wild-type Col as background) show a similar ABA insensitivity to ABI2-WT transgenic lines in ABA-induced inhibition of seed germination. Each value is the mean ±SE of five independent biological determinations. (D) ABI2-3A and ABI2-3D transgenic lines show a similar ABA insensitivity to ABI2-WT transgenic lines in early seedling growth. The seeds were directly planted in 3 μM ABA-containing medium and the growth status was recorded 12 d after stratification. The experiments were repeated five times with similar results.