Table 1.
The putative optimal codons and tRNA abundance
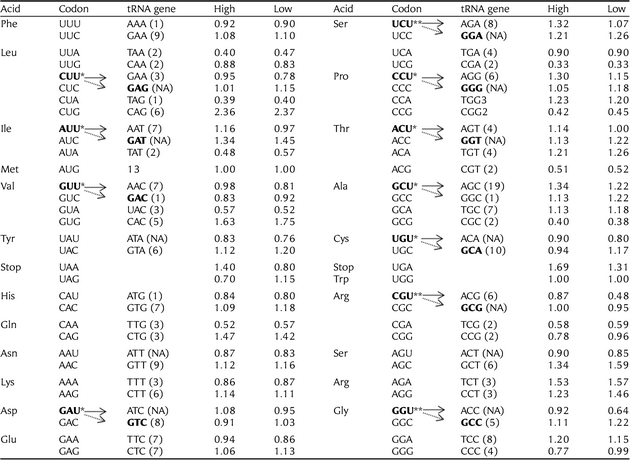 |
Putative optimal codons were inferred based on departures from equal codon usage by sets of loci with high (5% top) and low (5% down) gene expression (▵RSCU). ▵RSCU for a given codon is the difference between the average RSCU of genes with high and low expression (significance tested using the one-way ANOVA). If ▵RSCU is >0.1 at P< 0.05, this codon will be identified as optimal codon. Total optimal codons identified in this study are 11. The transfer RNA gene (tRNA) copy numbers for each codon was taken from http://gtrnadb.ucsc.edu/Ggall/. There is a good correspondence between tRNA abundance and optimal codons within codon classes. However, the above correlation reflects only partially the real co-adaptation of tRNA abundance and codon usage, as the same tRNA can decode several codons. Since we have no experimental data on base modifications in Gallus gallus tRNAs, we predicted the codons decoded by the different anticodons according to the ‘parsimony of wobbling’ criterion. The putative optimal codons with ▵RSCU is >0.1 at P< 0.05 and ▵RSCU >0.2 at P< 0.05 are denoted by ‘*’ and ‘**’, respectively.
