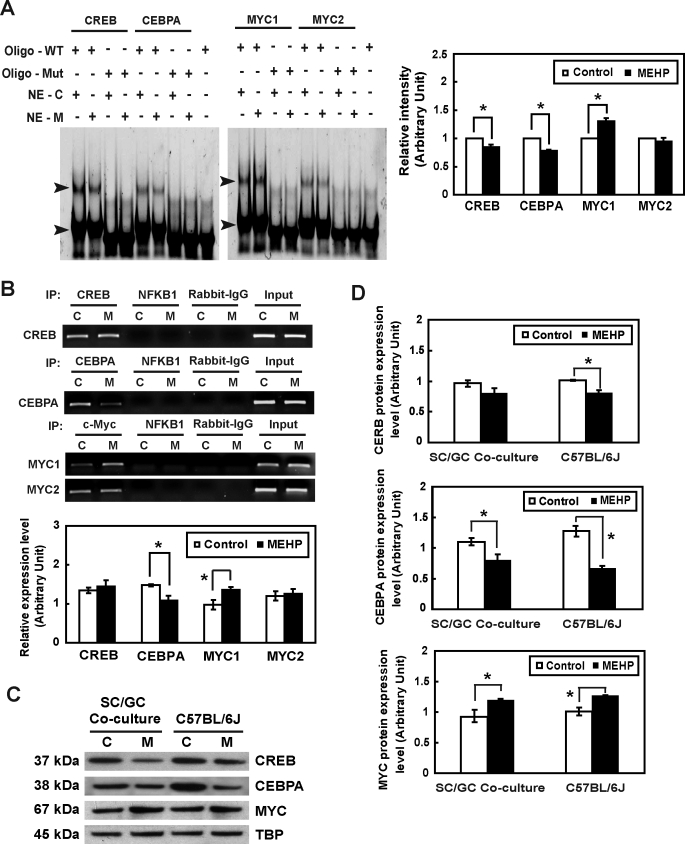
FIG. 4.
The influence of MEHP exposure on the transcriptional activity of Timp2 in vitro is shown. A) Detection of CREB, CEBPA, and MYC in Sertoli cells and their interactions with the Timp2 5′-UAS in response to MEHP by EMSA are shown. Nuclear extracts (NE) from untreated or MEHP-treated primary rat Sertoli cells were incubated with double-stranded oligonucleotides containing CREB, CEBPA, or MYC binding sequences specific to the Timp2 5′-UAS (oligo-WT) or with mutated oligonucleotides (oligo-Mut). The EMSA gel is stained with SYBR Green. The arrowhead indicates the oligonucleotide-protein complex. C, control; M, MEHP. Right) Quantitative data of shifted bands with high molecular weight are shown. Values represent means ±SEM with an asterisk denoting significant differences from control (*P < 0.05, Student t-test). B) Binding activities of CREB, CEBPA, and MYC to the Timp2 5′-UAS in response to MEHP were determined by ChIP assay. PCR was performed with DNA isolated from primary rat Sertoli cells and then subjected to immunoprecipitation with an anti-CREB, anti-CEBPA, or anti-MYC antibody. Specific primers to the Timp2 promoter amplified regions containing CREB, CEBPA, MYC1, and MYC2 binding sites. The input represents the amplification of the unprecipitated DNA. PCR amplification of irrelevant immunoprecipitation against anti-NFKB1 antibody and normal rabbit IgG were used as negative controls. Values represent means ± SEM, with the asterisk denoting significant differences from control (*P < 0.05, Student t-test). C) Nuclear extract (30 μg) from primary rat Sertoli cell-germ cell cocultures treated with 200 μM MEHP for 6 h and from C57BL/6J mice testes treated with 1 g/kg of MEHP for 6 h were analyzed by Western blot analysis using antibodies against MYC, CEBPA, and CREB. TBP served as the loading control. D) Quantitative data of Western blot analysis are shown. Values represent means ± SEM, with the asterisk denoting significant differences from control (*P < 0.05, Student t-test).