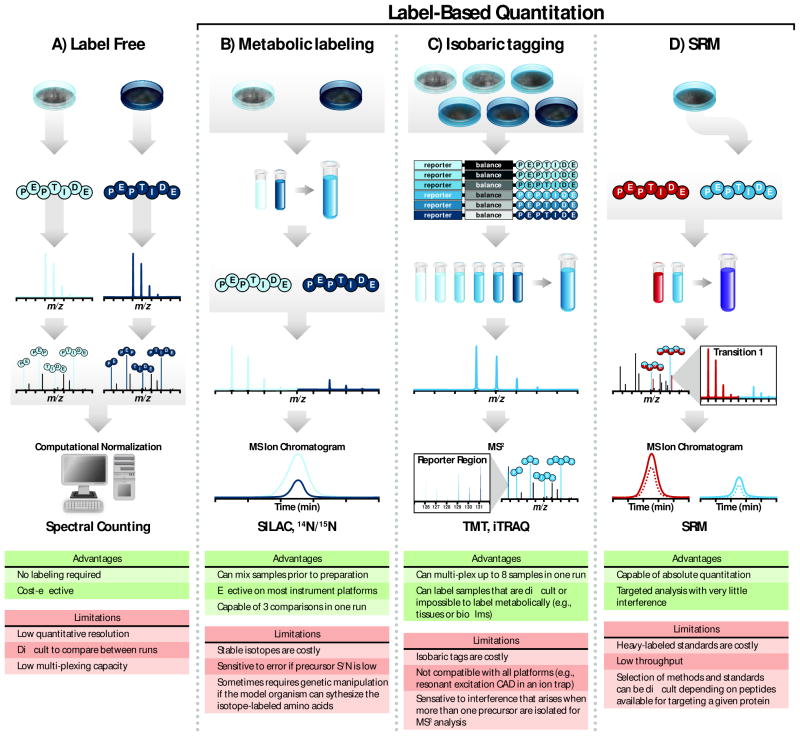
Figure 3.
Quantitative proteomics strategies. (A) During label-free quantitation, peptides extracted from samples are identified through tandem MS. Spectral counting measures the number of peptide spectral matches (i.e., the number of times the peptide is observed) followed by normalization that accounts for protein size. This technique allows for relative quantitation without the use of isotopes. (B) Metabolic labeling incorporates heavy isotopes into proteins through amino acids or nutrients (i.e., 15N or 13C). Differently-labeled samples are combined during protein preparation and their corresponding stable isotopes generate a shift in m/z values observed in MS1 scans. The difference between extracted ion chromatograms for peaks corresponding to heavy and light samples is proportional to the relative abundance of each. (C) Isobaric labeling strategies, such as TMT and iTRAQ, are capable of comparing up to six or eight samples in a single run depending on the method. Like metabolic labeling, samples are combined during protein preparation. Labeled peptides from different samples within the mixture have the same nominal mass and co-elute with reversed-phase chromatography. Peptide fragmentation during tandem mass spectrometry produces both sequence ions for peptide identification and reporter tags for quantitation. (D) Quantitation via single reaction monitoring (SRM) is capable of absolute quantitation and is often performed on triple quadruple mass spectrometers. Peptides from samples are mixed with a chemically synthesized heavy peptide that serves as a quantitation standard. Because the exact amount of this standard is known, absolute quantitation is possible through this method. Figure panels adapted from (Gerber et al., 2007) and (Ross et al., 2004). Color figures are available online.