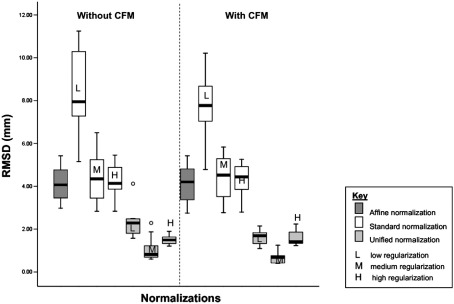
Fig. 5.
Experiment 2: Normalization of lesioned brains with and without CFM. Plot of the root mean squared difference (RMSD) values in millimeters for the 14 normalizations of the group of ten simulated lesion–normal brain pairs. From the left of the image the first (dark gray) box-plot shows RMS values for the affine-only solution (mean—4.1, s.d—0.86), then next the three (white) standard solutions with low (L) (mean—8.36, s.d—1.82), medium (M) (mean—4.4, s.d—1.12) and high (H) (mean—4.31, s.d—0.77) regularizations. The subsequent 3 box-plots in light gray show RMS values for unified solutions with low (L) (mean—2.33, s.d—0.71), medium (M) (mean—1.09, s.d—0.58) and high (H) (mean—1.52, s.d—0.25) regularizations. The 7 subsequent box-plots, after the dotted line with shaded background, show the same normalization solutions with cost function masking (CFM): affine (mean—4.13, s.d—0.88), standard solutions with low (L) (mean—7.82, s.d—1.54), medium (M) (mean—4.35, s.d—1.07) and high (H) (mean—4.35, s.d—0.77) regularizations, unified solutions with low (L) (mean—1.62, s.d—0.32), medium (M) (mean—0.72, s.d—0.31) and high (H) (mean—1.57, s.d—0.34) regularizations. The black line in each box-plot indicates the group mean for each normalization solution. The open circle indicates a group outlier.