Abstract
Carotenoid-based coloration has attracted much attention in evolutionary biology owing to its role in honest, condition-dependent signalling. Knowledge of the genetic pathways that regulate carotenoid coloration is crucial for an understanding of any trade-offs involved. We identified genes with potential roles in carotenoid coloration in vertebrates via (i) carotenoid uptake (SR-BI, CD36), (ii) binding and deposition (StAR1, MLN64, StAR4, StAR5, APOD, PLIN, GSTA2), and (iii) breakdown (BCO2, BCMO1). We examined the expression of these candidate loci in carotenoid-coloured tissues and several control tissues of the red-billed quelea (Quelea quelea), a species that exhibits a male breeding plumage colour polymorphism and sexually dimorphic variation in bill colour. All of the candidate genes except StAR1 were expressed in both the plumage and bill of queleas, indicating a potential role in carotenoid coloration in the quelea. However, no differences in the relative expression of any of the genes were found among the quelea carotenoid phenotypes, suggesting that other genes control the polymorphic and sexually dimorphic variation in carotenoid coloration observed in this species. Our identification of a number of potential carotenoid genes in different functional categories provides a critical starting point for future work on carotenoid colour regulation in vertebrate taxa.
Keywords: carotenoid coloration, candidate genes, condition-dependent signalling, evolutionary biology
1. Introduction
Animal coloration has long been used as a model for investigating adaptive evolution in wild populations [1]. Carotenoid-based coloration, in particular, has become a focus of many studies in behavioural and evolutionary ecology, because of its important role in condition-dependent signalling [2]. Carotenoids are bright yellow and red pigments that are responsible for some of the most conspicuous coloration found in vertebrates. However, there are a number of factors that potentially limit the ability of animals to deposit carotenoids in their integument. Unlike other pigment types such as melanins, carotenoids cannot be synthesized by vertebrates and must instead be acquired through the diet [3]. There are potential costs associated with the acquisition of carotenoids and their transport and metabolism within the body [4], as well as hypothesized trade-offs between their uses in coloration and other physiological roles, such as acting as antioxidants in the immune system [5]. This means that carotenoid-based colour traits, when carotenoids are limited in availability, are potentially costly to produce and maintain, and so could function as honest, reliable signals of quality to prospective mates or dominance to rivals. A number of studies have provided support for this hypothesis in a range of wild species with carotenoid-based traits [2,4].
Although carotenoid traits have often been shown to be condition-dependent, carotenoid coloration is also dependent on underlying genetic mechanisms. Birds selectively accumulate carotenoids in various parts of their integument, preferentially accumulate certain carotenoids over others, and are able to enzymatically convert dietary carotenoids into more colourful derived forms [2]. This level of physiological control of carotenoid coloration strongly implies the involvement of appropriate carotenoid-binding and transport proteins and enzymes along with the genes that encode them [6]. Knowledge of the genetics underlying carotenoid colour variation is required in order to provide a mechanistic understanding of the potential costs involved in carotenoid signalling and how sexual selection operates at a molecular level. Moreover, it is of considerable interest to know whether similar molecular mechanisms are used for the carotenoid pigmentation of different tissues both within and among species as this could potentially lead to an insight into convergent evolution at the molecular level, or alternatively reveal a range of different mechanisms that birds use to achieve similar phenotypic outcomes. Knowledge of the genetics of carotenoid-based signals would also help to understand the potential trade-offs between carotenoids used in displays and those used in other physiological roles. However, to date there has been almost no progress made in determining the genetic basis of carotenoid coloration in vertebrates. A major reason for this is the lack of suitable vertebrate model systems for resolving the carotenoid pathway.
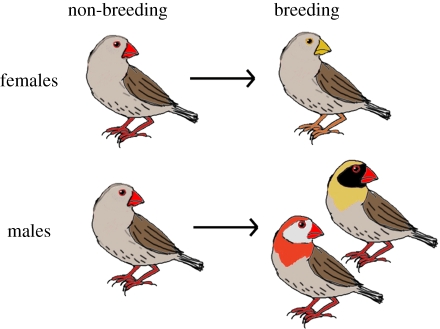
The red-billed quelea (Quelea quelea) provides an excellent opportunity to investigate the genetic basis of carotenoid-based colour variation in a wild vertebrate. This species is a socially monogamous, seed-eating passerine that is a major crop-pest in sub-Saharan African countries [7]. It is one of the most abundant species of bird in the world, and it is also one of the most variable species in terms of its coloration, both within and between sexes (figure 1). Throughout their non-breeding season, queleas are sexually monomorphic, with a melanin-based dull brown plumage, and a carotenoid-based red bill, eye-rings and legs [8–10]. The red coloration in bills and feathers is due to the presence of enzymatically derived keto-carotenoids similar to those found in several other bird species, such as the zebra finch ([8,9], table 1). Prior to the breeding season, female queleas lose the carotenoid coloration of their bare body parts. Breeding males, however, retain this coloration and also moult into a bright nuptial plumage that is polymorphic for both breast colour that varies from red to buff and for face colour that varies from white to black (figure 1). The breast colour polymorphism is due to differences in carotenoid deposition and is inherited in a quasi-Mendelian fashion [8,9]. In buff-breasted males, carotenoids are absent, the buff coloration being owing to phaeomelanin, whereas red-breasted males have variable amounts of keto-carotenoids present. The mask coloration is due to differential levels of eumelanin deposition and also has a strong genetic basis. The mask and breast polymorphisms assort independently of each other [8,9] and both traits are developmentally fixed, genetically determined characters of which the expression appears to be independent of phenotypic condition or environmental variation [8,9]. The polymorphic breeding plumage is hypothesized to be under sexual selection for signalling individual identity among neighbouring territorial males and the high levels of variability present are probably maintained through negative frequency-dependent selection [11,12]. In contrast, the red bill of the quelea, as well as the other bare body parts, appears to be a condition-dependent indicator of quality [8,9]. In breeding quelea males, the redness of the bill correlates with overall body condition [8,9] as well as with social dominance [13], effectively serving as a badge of status.
Figure 1.
Plumage and bill colour variation in the red-billed quelea. Throughout their non-breeding season, queleas are sexually monomorphic. Female queleas lose the carotenoid coloration of their bare body parts prior to the breeding season, while breeding males retain this coloration and also moult into a bright nuptial plumage that is polymorphic for both breast colour that varies from red to buff and for face colour that varies from white to black. Flank feathers, used here as controls, never contain carotenoids in any male plumage.
Table 1.
Concentrations (µg pigment/g feather) and relative percentages of carotenoids recovered from the plumage and bill of red-billed queleas.
| male no. | astaxanthin | α-doradexanthin | adonirubin | canthaxanthin | |
|---|---|---|---|---|---|
| male red plumage | 1 | 16.8 (59.4%) | 6.5 (22.8%) | 4.4 (15.4%) | 0.7 (2.4%) |
| 2 | 26.8 (57.2%) | 11.2 (23.8%) | 8.1 (17.2%) | 0.8 (1.8) | |
| 3 | 30.7 (59.6%) | 12.4 (24.0) | 7.3 (14.1%) | 1.1 (2.2%) | |
| male buff plumage | 2a | —(0%) | — (0%) | — (0%) | — (0%) |
| 4 | — (0%) | — (0%) | — (0%) | — (0%) | |
| 5 | — (0%) | — (0%) | — (0%) | — (0%) | |
| male red billb | pooledc | (56.9%) | (26.6%) | (13.1%) | (3.3%) |
| female yellow bill | pooledc | (0%) | (0%) | (0%) | (0%) |
aThis male had a red breast and a buff crown, and so is included in both plumage groups.
bNote that absolute concentrations of carotenoids in the bill were not determined (see §2).
cPigments extracted from bills were extracted from the pooled samples of five individuals.
The genetic mechanism involved in the carotenoid-based plumage polymorphism of male queleas is likely to involve a difference in expression of one or a few genes at the site of deposition in the feather follicles. Genes involved in determining carotenoid coloration are likely to be pleiotropic, influencing carotenoid metabolism throughout the body, and coding mutations in these genes are predicted to result in broad phenotypic effects. Therefore, regulatory mutations are more likely to be responsible for the quelea polymorphism, as these could up- or downregulate the expression of the genes at the integument without disrupting their other functions in the body. Buff-plumage morphs, like red-plumage morphs, express carotenoid-based red coloration in their bare body parts, and circulate these same carotenoids in their serum as well as lutein and zeaxanthin [14]. This indicates that buff-plumage morph males retain the global ability to acquire, metabolize and transport carotenoids, and that the expression difference responsible for the plumage polymorphism is tissue-specific, taking place at the site of deposition. This makes quelea carotenoid plumage polymorphism an ideal model for an expression study investigating the differential expression of candidate genes that are involved in carotenoid coloration. Moreover, the genetic control of sexual dimorphism in bill colour provides an intriguing contrast to the male plumage polymorphism since here the regulation is sex-specific and involves a bare body part rather than feathers. In this case, part of the regulatory mechanism probably involves differences in circulating carotenoids between the sexes, in addition to gene regulation in the bill. Although the ultimate mechanism (genetic polymorphism versus sex-specific regulation) is different, an important question is whether the downstream genetic pathways controlling differential deposition of carotenoids are shared.
The identification of candidate genes for carotenoid deposition in birds relies on (i) biochemical and physiological studies on carotenoid uptake, metabolism and deposition in vertebrates [6,15,16], (ii) recent work on the genetics of carotenoid coloration in chicken legs [17], and (iii) studies on carotenoid deposition in arthropods [18–20]. From these sources, we compiled a series of 11 carotenoid candidate genes (table 3). For a detailed description of these loci and their potential relevance to carotenoid coloration, see the electronic supplementary material.
Table 3.
Relative expression of candidate genes when compared with β-actin. Mean expression levels and standard deviations for each set of biological replicates are shown. n.d., not determined. *p < 0.05, **p < 0.01.
| gene | red plumage (n = 3) | buff plumage (n = 3) | flank plumage (n = 3) | red (n = 3) versus buff (n = 3) p-value | male bill (n = 3) | female bill (n = 3) | male bill (n = 3) versus female bill (n = 3) p-value | plumage (n = 9) versus bill (n = 6) p-value | liver (n = 3) | duodenum (n = 3) | plumage (n = 9) versus liver (n = 3) p-value | plumage (n = 9) versus duodenum (n = 3) p-value | bill (n = 6) versus liver (n = 3) p-value | bill (n = 6) versus duodenum (n = 3) p-value |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BCO2 | 0.03923 ± 0.03331 | 0.00874 ± 0.00211 | 0.00251 ± 0.00133 | 0.25 | 0.00006 ± 0.00001 | 0.00004 ± 0.00003 | 0.27 | 0.07 | 0.00001 ± 0.000004 | 0.000003 ± 0.000002 | 0.07 | 0.07 | 0.008** | 0.006** |
| BCMO1 | 0.00031 ± 0.00008 | 0.00034 ± 0.00026 | 0.00033 ± 0.00026 | 0.86 | 0.00057 ± 0.00037 | 0.00026 ± 0.00009 | 0.27 | 0.52 | 0.00859 ± 0.00740 | 0.06553 ± 0.05574 | 0.19 | 0.18 | 0.20 | 0.18 |
| SR-BI | 0.00403 ± 0.00325 | 0.00264 ± 0.00276 | 0.00049 ± 0.00011 | 0.60 | 0.04092 ± 0.00316 | 0.02975 ± 0.01454 | 0.31 | 0.0007** | 0.09297 ± 0.07052 | 0.00618 ± 0.00230 | 0.16 | 0.08 | 0.29 | 0.001** |
| CD36 | 0.05678 ± 0.03337 | 0.13037 ± 0.06995 | 0.08603 ± 0.06034 | 0.20 | 0.02938 ± 0.02282 | 0.01727 ± 0.00933 | 0.46 | 0.009** | 3.03904 ± 3.61723 | 0.38498 ± 0.35273 | 0.29 | 0.29 | 0.29 | 0.22 |
| MLN64 | 0.01628 ± 0.01242 | 0.01382 ± 0.01272 | 0.00318 ± 0.00351 | 0.82 | 0.01395 ± 0.00830 | 0.00719 ± 0.00405 | 0.30 | 0.91 | 0.02134 ± 0.02730 | 0.00430 ± 0.00596 | 0.59 | 0.22 | 0.57 | 0.22 |
| StAR4 | 0.07981 ± 0.02448 | 0.10387 ± 0.03967 | 0.11402 ± 0.06960 | 0.43 | 0.08149 ± 0.04111 | 0.16242 ± 0.08455 | 0.24 | 0.52 | 0.61949 ± 0.22698 | 0.12981 ± 0.03250 | 0.06 | 0.26 | 0.06 | 0.83 |
| StAR5 | 0.00251 ± 0.00074 | 0.00337 ± 0.00229 | 0.00284 ± 0.00046 | 0.59 | 0.00288 ± 0.00108 | 0.00275 ± 0.00058 | 0.87 | 0.87 | 0.01940 ± 0.01419 | 0.00293 ± 0.00136 | 0.18 | 0.98 | 0.18 | 0.90 |
| APOD | 0.01404 ± 0.01108 | 0.02488 ± 0.02358 | 0.00350 ± 0.00052 | 0.53 | 0.78296 ± 0.94122 | 1.13758 ± 0.59894 | 0.62 | 0.02* | 0.08145 ± 0.12270 | 0.00037 ± 0.00015 | 0.44 | 0.03* | 0.03* | 0.02* |
| PLIN | 0.00046 ± 0.00028 | 0.00069 ± 0.00030 | 0.00196 ± 0.00105 | 0.38 | 0.03209 ± 0.05374 | 0.00280 ± 0.00391 | 0.44 | 0.33 | 0.00551 ± 0.00818 | 0.08293 ± 0.07251 | 0.14 | 0.19 | 0.49 | 0.25 |
| GSTA2 | 0.03536 ± 0.02087 | 0.05886 ± 0.01246 | 0.03188 ± 0.02073 | 0.19 | 0.00432 ± 0.00218 | 0.00462 ± 0.00158 | 0.86 | 0.0005** | 0.52517 ± 0.48926 | 0.01798 ± 0.01259 | 0.23 | 0.053 | 0.21 | 0.20 |
| PKCIW | 0 ± 0 | 0 ± 0 | n.d. | n.d. | 0 ± 0 | 0.52498 ± 0.14334 | 0.02* | 0.09 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
In this study, we first determined the specific carotenoids present in the bills and feathers of queleas in breeding condition. Second, we quantified the expression of the 11 identified candidate genes in tissues actively depositing carotenoids in the quelea, and investigated whether differential expression was associated with colour variation in feathers of buff versus red-morph males, and bills of males versus females. We also quantified the expression of these candidate genes in feathers taken from the flank region of male queleas where carotenoid deposition does not occur, in order to provide an additional control for the experiment, and also in the duodenum and the liver of male queleas, which have important roles in the uptake and metabolism of carotenoids, respectively.
2. Material and methods
(a). Determination of carotenoids in tissues
Plumage samples from five males in breeding condition were collected from a breeding colony in Zimbabwe. Two of these males were red morphs and two were buff morphs, while one male had a red-morph breast and a buff-morph crown. The feathers were divided into three samples each of red and buff, respectively. Carotenoids were extracted using a traditional acidified-pyridine chemical technique [39] and were analysed following the high-performance chromatographic methods of McGraw et al. [40]. Authentic reference carotenoids were provided by R. Stradi and were run as external standards. Shavings from the bills of five males (red bills) and five females (yellow bills) were pooled separately and analysed as above. However, for bills, carotenoid concentrations were not determined because the pigments were esterified (as in zebra finch bills; [41]) and thus we could not confidently resolve absolute amounts of esterified peaks with this analytical system.
(b). Samples
Quelea feather tissue samples were collected in September 2009 from a field site in the Orange River region of central South Africa. Feather samples were plucked from the breast and flank region of male queleas that were in the process of moulting into breeding plumage, at an early stage corresponding to the period of carotenoid deposition. Quelea bill tissue samples were taken from breeding males and females and quelea liver and duodenal tissue samples were taken from breeding males collected in March 2009 in Zimbabwe. Shavings of bill tissue were taken from the rhamphotheca, where carotenoid deposition occurs. All samples were stored in RNAlater solution (Ambion).
(c). Laboratory methods
Sequence alignments were constructed from the gene sequences of zebra finch and chicken, obtained from GenBank, and used to design intron-spanning primers for the candidate genes in quelea using Primer3Plus (http://www.bioinformatics.nl/primer3plus). Total RNA was extracted from the avian tissue samples using the RNeasy Mini Kit (Qiagen) and treated with Deoxyribonuclease I, Amplification Grade (Invitrogen) to remove residual genomic DNA carry over. Complementary DNA (cDNA) was prepared from total RNA using Superscript II Reverse Transcriptase (Invitrogen). Quantitative real-time RT-PCR was performed for each candidate locus on cDNA from breast plumage of red (n = 3) and buff (n = 3) male-morph individuals, bill tissue of male (n = 3) and female (n = 3) individuals, and flank plumage, liver and duodenal tissues of male individuals (n = 3). In the case of plumage samples, total RNA extracted from 5–15 feather follicles was pooled per individual. Reactions were performed using SYBR Green (Qiagen) in an Opticon-2 PCR machine (MJ Research). The housekeeping gene β-actin was used for normalization. A female-specific locus, protein kinase C inhibitor (PKCIW), was used as a positive control. Three technical replicates were used for each biological sample (see the electronic supplementary material for further experimental details). Tests for statistical significance were performed using unpaired two-tailed t-tests assuming unequal variance.
3. Results
(a). Carotenoid expression in tissues
Plumage samples from three separate red-morph males recovered four ketocarotenoid types (primarily astaxanthin and alpha doradexanthin, table 1). These carotenoids were expressed at slightly different concentrations between the different individuals, but at similar relative ratios. By contrast, plumage from buff males (n = 3) recovered no carotenoids. The carotenoids in male bills were the same four present in red plumage, and expressed at remarkably similar ratios (table 1). None of these ketocarotenoid pigments are known to be present in the diets of granivorous songbirds like queleas [42], which eat primarily wild grass seed, millet, sorghum and wheat, so we presume that the red coloration of plumage and bill coloration very likely results from the outcome of the same metabolic conversions of dietary xanthophylls (also see [43] in common crossbills, Loxia curvirostra). No carotenoids were present in the (yellow) bills of breeding females.
(b). Expression of candidate genes in quelea tissue samples
Quantitative real-time reverse transcription PCR was performed for each of the 11 candidate genes (table 2) to determine their relative levels of expression in developing feather follicles taken from the breast region of male red and buff-morph queleas and the flank region of male queleas (where carotenoids are never deposited), bill tissue taken from male and female breeding individuals, and liver and duodenal tissues taken from breeding male individuals. All of the candidate loci, with the exception of StAR1, were expressed in all of the tissue samples tested (table 3). As a positive control for StAR1, reactions were performed using quelea ovary cDNA, a tissue in which StAR1 is known to be expressed in mammals [44]. Expression in this tissue indicates that the primers used were suitable for amplification of StAR1 in quelea, but that this gene is not expressed in the plumage, bill, liver or duodenum. The results of tests for expression differences in seven separate class comparisons are shown in table 2: red-morph plumage versus buff-morph plumage, male bill versus female bill, plumage versus bill, plumage versus liver, plumage versus duodenum, bill versus liver and bill versus duodenum. As expected, the female-specific locus PKCIW was expressed in the female bill, but not in the male tissues, and this difference was significant (table 3). Comparison of the expression patterns of the remaining genes revealed no difference in expression of any of the genes between the red and buff morphs or between male and female bills (table 3). Additionally, there was no significant difference in expression of any of the genes between the flank plumage and the breast plumage (not shown), so the three plumage categories and two bill categories were pooled for further analyses among tissues. Four candidate genes were differentially expressed between the plumage and bill: SR-BI and APOD were more highly expressed in the bill, whereas CD36 and GSTA2 were more highly expressed in plumage (table 3). APOD was more highly expressed in the plumage compared with the duodenum and APOD was also more highly expressed in the bill compared with the liver and duodenum (table 3). SR-BI was more highly expressed in the bill compared with the duodenum and BCO2 was more highly expressed in the bill compared with the liver and duodenum (table 3).
Table 2.
Candidate genes for carotenoid coloration in the red-billed quelea.
| name | known function | evidence for potential role in carotenoid coloration | key references |
|---|---|---|---|
| BCO2(formerly BCDO2) | asymmetrical cleavage of carotenoids to form precursors of vitamin A and other metabolites | responsible for a carotenoid polymorphism in chicken skin; lower expression levels lead to the retention of carotenoids and a yellow skin phenotype | [17,21] |
| BCMO1 | symmetrical cleavage of carotenoids to form vitamin A; believed to be specific to β-carotene | similar function in carotenoid breakdown to BCO2 | [22,23] |
| SR-BI | membrane-bound lipid and carotenoid transporter | involved in the uptake of carotenoids in intestine and retina; homologous to carotenoid-binding gene ninaD in Drosophila | [19,24] |
| CD36 | membrane-bound lipid and carotenoid transporter | possible role in carotenoid binding in retina; homologous to carotenoid-binding gene Cameo2 in Bombyx mori | [20,25] |
| StAR1 | intracellular cholesterol transporter for steroidogenesis | similar to B. mori carotenoid-binding protein (CBP); closely related to MLN64 | [26–28] |
| MLN64 | intracellular transporter of cholesterol and other lipids | orthologue of Bombyx CBP | [26,27,29] |
| StAR4 | intracellular transporter of lipids; related to other StAR genes | possible candidate for CBP human retina lutein binding protein (HR-LBP) in human retina | [30,31] |
| StAR5 | intracellular transporter of lipids; related to other StAR genes | possible candidate for CBP HR-LBP in human retina | [30,31] |
| APOD | multi-functional, multi-ligand component of lipoproteins | related to CBP crustacyanin in the carapace of crustaceans; expressed in bird feathers | [32,33] |
| PLIN | involved in packaging or breakdown of lipid droplets within cells of the adipose tissue | the likely association of carotenoids with lipid droplets within cells suggests the possible involvement of PLIN | [34,35] |
| GSTA2 | conjugates electrophillic compounds to glutathione; and involved in detoxification and transport | the closest avian homologue of the carotenoid-binding gene GSTP1 in the mammalian retina; also expressed in duodenum from where carotenoids are selectively uptaken | [36–38] |
4. Discussion
The determination of carotenoid concentrations in feather and bill samples of red-billed queleas confirmed that there are large differences in the concentration of four ketocarotenoids among feathers from red- and buff-morph males, and showed that the same four ketocarotenoids are present in similar ratios in male bills and absent from female bills. Although there is little known about the genes involved in carotenoid coloration in vertebrates, our review of the literature highlighted several plausible candidates in a range of functional categories, including carotenoid uptake (SR-BI, CD36), carotenoid binding and deposition (StAR1, MLN64, StAR4, StAR5, APOD, PLIN, GSTA2) and carotenoid breakdown (BCO2, BCMO1). Ten out of the 11 candidate genes were expressed in both developing feathers and bill tissue of both sexes in queleas. However, there were no detectable differences in the expression of any of the candidate genes between red and buff male quelea-morph feathers or between male and female bill. Thus, it is very likely that the regulation of male-morph coloration and sexually dimorphic bill coloration is ultimately controlled by other loci. In addition, the expression of the loci in tissues where carotenoids are not deposited (buff-morph male breast feathers, and male flank feathers) suggests that the loci have a function unrelated to carotenoids, that may in some cases reflect a broader role in lipid metabolism. Nevertheless, as we discuss below, an additional function of the loci in carotenoid coloration cannot be ruled out, especially given the striking evolutionary conservation emerging in the function of some gene families that are involved in carotenoid metabolism.
The lack of differential expression of any of the candidate genes between the red and buff morphs or between male and female bills indicates that expression differences in the genes are unlikely to be responsible for these differences in carotenoid coloration in the quelea. We were able to detect significant expression differences for a control locus (PKCIW) but the relatively small sample sizes used mean that minor expression differences between phenotypes would be difficult to detect statistically. We were careful to sample tissues at the time of carotenoid deposition so the negative result cannot be attributed to sampling at the wrong time.
It is important to consider the possibility that mechanisms other than expression differences in the tissue where carotenoid deposition is occurring may be responsible for the variation in carotenoid coloration. There have been cases where variation in the coding region of BCO2 has been associated with carotenoid variation in non-integumentary tissues of domesticated mammals. Nonsense mutations in BCO2 are responsible for the accumulation of carotenoids within adipose tissue of Norwegian sheep [45], and in the milk of cows [46], giving a yellow coloration to the fat and milk, respectively. However, protein-coding variation in genes involved in carotenoid coloration is likely to have significant pleiotropic effects, given their roles in other areas of lipid metabolism. Since there are no phenotypic differences other than plumage coloration apparent among the red and buff morphs, gene expression differences at the site of carotenoid deposition remain the most probable cause of carotenoid variation in the male plumage polymorphism. The situation is different for variation in bill colour, since the downregulation of carotenoid coloration in the breeding female quelea bare body parts may be largely owing to physiological changes in the body, rather than the differential expression of genes at the site of deposition in the integument. When in breeding condition, yellow-billed female queleas have sharply lower serum carotenoid concentrations than both males or non-breeding females (J. Dale 2011, unpublished data) and this is presumably directly related to the loss of carotenoid coloration in their bare body parts.
The lack of differential expression of BCO2 in the plumage of red and buff quelea morphs indicates that the quelea plumage polymorphism is probably under different genetic control to that found in chicken skin, where expression differences in BCO2 are responsible for the differential cleavage of carotenoids between white and yellow skinned chickens. Birds use different strategies to produce their colourful carotenoid traits and these may reflect differences in the genetic mechanisms employed. For example, bishop weavers (Euplectes spp.) produce red plumage through the metabolism of yellow dietary carotenoids to derived keto-carotenoids, whereas the closely related widowbirds achieve red coloration through the deposition of very high concentrations of yellow dietary carotenoids [47]. Some birds, such as the house finch, Carpodacus mexicanus, achieve a brighter coloration through the accumulation of large amounts of a variety of carotenoid types [48] whereas other species, such as the American goldfinch, Carduelis tristis, use the selective deposition of specific carotenoids to achieve their bright coloration [49]. These variable strategies may be the result of differences in the complement of carotenoid-binding proteins and metabolic enzymes that birds are able to employ to produce their coloration, and this may affect the signal content of their carotenoid displays.
Although the loci examined here are unlikely to be responsible for ultimately controlling carotenoid colour variation in queleas, a role of some of them in carotenoid deposition remains likely. One remarkable feature of the carotenoid pathway in animals highlighted by our literature review is the high level of conservation that exists among taxa, both at the physiological and genetic level. This probably reflects the similarities in general lipid metabolic pathways that exist between animal groups, such as mammals and insects, as well as a number of homologues of lipid-binding genes that are shared among these taxa [50]. It is interesting to note that this is in contrast to the situation for melanin-based pigmentation, which involves different genetic mechanisms in vertebrates and invertebrates [1,51]. The conservation of carotenoid metabolism may extend to interactions among multiple proteins at different points in the pathway. Most notably, as seen in the mammalian eye, silkworm cocoon and other examples, members of the scavenger receptor family (e.g. SR-BI/CD36) are involved in carotenoid uptake into cells while members of the START family are involved in carotenoid deposition [52,53]. Thus, a role of members of these families in carotenoid coloration in queleas remains likely.
An interesting issue is whether carotenoid coloration of different integumentary parts within an individual may be under different genetic control. Since the carotenoid coloration of the plumage and bare body parts of the quelea appear to be under different selective pressures [8,9], it is possible that there may be different genes involved in the production of these traits. It was notable, therefore, that the two scavenger receptor genes showed significant differences in the expression level between the two tissues, with SR-BI (as well as APOD) more highly expressed in the bill, whereas CD36 (as well as GSTA2) was more highly expressed in plumage. These differences in expression raise the possibility that these genes may play different roles in the production of carotenoid coloration in different tissues. Nevertheless, the similar ratios of ketocarotenoids in red-morph feathers and male bills suggest some commonality in the biochemical mechanisms of deposition in the two tissue types.
The expression of all of the candidate genes, with the exception of StAR, in the quelea duodenum and liver was as expected in line with their important role in carotenoid uptake and metabolism. However, there were some interesting differences between the expression patterns of these genes in the integumentary organs compared with the liver and duodenum. BCO2 was more highly expressed in the bill than in either the liver or duodenum. The relatively high expression of this carotenoid-cleavage enzyme in a tissue, where carotenoids are deposited at a high concentration is unexpected, as is its low expression in the liver and duodenum, which are major storage and production sites for vitamin A in the body [54,55]. It is worth noting that the mean expression level of BCO2 in the plumage was even higher than in the bill, although this was not significantly different owing to high levels of variability among individuals. Also of interest is the higher expression of SR-BI in the bill compared with the duodenum, since SR-BI is important for the uptake of carotenoids and other lipids from the intestine [56]. Finally, the higher expression of APOD in the plumage compared with the duodenum and in the bill compared with the liver and duodenum fits with the expected pattern of expression of this gene, as APOD is expressed highly in developing chicken feathers and skin, but at low levels in the liver [32].
The candidate gene approach is limited by the fact that effective choice of candidate loci depends on having prior knowledge of the possible functions and roles of the candidates. As such there is a need for alternative experimental approaches to the problem of identifying carotenoid coloration genes. Association and linkage studies could be used to determine quantitative trait loci across the whole genome associated with carotenoid variation. This approach is generally difficult to apply to wild populations such as the quelea. However, the zebra finch (Taeniopygia guttata) is another passerine that exhibits carotenoid coloration in its bill, and the recent sequencing of the zebra finch genome and the availability of zebra finch breeding lines would enable whole genome scans to be conducted in this species.
To conclude, we have conducted the first detailed study into the genetic basis of naturally occurring carotenoid-based colour variation in a wild vertebrate species. Most of the genes identified as candidates for a possible role in carotenoid coloration in the red-billed quelea were expressed in both developing feathers and bill. However, no association was found between the level of expression of the candidate genes and variable expression of carotenoid coloration. This indicates that the carotenoid breast-colour polymorphism present in red-billed queleas is likely due to a different mechanism than the yellow/white skin polymorphism present in chickens, and that variation in other genes is probably responsible for determining the quelea plumage polymorphism. Additionally, there was no evidence found to indicate the involvement of these genes in the underlying gender-specific differences in carotenoid coloration in red-billed queleas. It is interesting that male and female individuals have such similar expression patterns of the candidate genes considering that each sex would be expected to have different patterns in the allocation of carotenoid resources during the breeding season. Although the genetic basis of carotenoid variation in the quelea integument was not identified, it is likely that some of the candidate genes investigated in this study have a role in the production of carotenoid coloration in the quelea and other vertebrates on the basis of their carotenoid binding roles in other tissues and species. In particular, the evolutionary conservation of a role of SR-BI/CD36 and START gene family members in carotenoid metabolism across vertebrates and invertebrates strongly suggests their likely involvement in carotenoid coloration. The identification of these potential carotenoid genes represents a well-grounded starting point for future work into the genetics of carotenoid coloration across all vertebrate taxa.
Acknowledgements
We would like to thank Martie and Attie Stander for generous assistance while at the South African field site, and Bruce Clegg, Sarah Clegg, Colin Wynham and the staff at Malilangwe Wildlife Reserve for assistance at the Zimbabwean field site. We thank several anonymous reviewers for comments which substantially improved the paper, and BBSRC, Leverhulme Trust and Max Planck Institute (Germany) for funding.
References
- 1.Hoekstra H. E. 2006. Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 97, 222–234 10.1038/sj.hdy.6800861 (doi:10.1038/sj.hdy.6800861) [DOI] [PubMed] [Google Scholar]
- 2.Hill G. E., McGraw K. J. 2006. Bird coloration. Volume 2: function and evolution. Cambridge, UK: Harvard University Press [Google Scholar]
- 3.Goodwin T. W. 1984. The biochemistry of the carotenoids. London, UK: Chapman and Hall [Google Scholar]
- 4.Olson V. A., Owens I. P. F. 1998. Costly sexual signals: are carotenoids rare, risky or required? Trends Ecol. Evol. 13, 510–514 10.1016/S0169-5347(98)01484-0 (doi:10.1016/S0169-5347(98)01484-0) [DOI] [PubMed] [Google Scholar]
- 5.Von Schantz T., Bensch S., Grahn M., Hasselquist D., Wittzell H. 1999. Good genes, oxidative stress and condition-dependant sexual signals. Proc. R. Soc. Lond. B 266, 1–12 10.1098/rspb.1999.0597 (doi:10.1098/rspb.1999.0597) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brush A. H. 1990. Metabolism of carotenoid pigments in birds. Faseb J. 12, 2969–2977 [DOI] [PubMed] [Google Scholar]
- 7.Bruggers R. L., Elliott C. C. H. 1989. Quelea quelea: Africa's bird pest. Oxford, UK: Oxford University Press [Google Scholar]
- 8.Dale J. 2000. Functional significance of ornamental plumage in red-billed queleas Quelea quelea. PhD thesis Cornell University, Ithaca, NY [Google Scholar]
- 9.Dale J. 2000. Ornamental plumage does not signal male quality in red-bill queleas. Proc. R. Soc. Lond. B 267, 2143–2149 10.1098/rspb.2000.1261 (doi:10.1098/rspb.2000.1261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward P. 1966. Distribution, systematics, and polymorphism of the African weaver-bird Quelea quelea. Ibis 108, 34–40 10.1111/j.1474-919X.1966.tb07250.x (doi:10.1111/j.1474-919X.1966.tb07250.x) [DOI] [Google Scholar]
- 11.Dale J., Lank D. B., Reeve H. K. 2001. Signaling individual identity vs. quality: a model and case studies with ruffs, queleas, and house finches. Am. Nat. 158, 75–86 10.1086/320861 (doi:10.1086/320861) [DOI] [PubMed] [Google Scholar]
- 12.Tibbetts E. A., Dale J. 2007. Individual recognition: it is good to be different. Trends Ecol. Evol. 22, 529–537 10.1016/j.tree.2007.09.001 (doi:10.1016/j.tree.2007.09.001) [DOI] [PubMed] [Google Scholar]
- 13.Shawcross J. E., Slater P. J. B. 1984. Agonistic experience and individual recognition in male Quelea quelea. Behav. Process. 9, 49–60 10.1016/0376-6357(84)90007-X (doi:10.1016/0376-6357(84)90007-X) [DOI] [PubMed] [Google Scholar]
- 14.Laucht S., Kempenaers B., McGraw K. J., Arriero E., Goymann W., Hasselquist D., Dale J. Submitted No evidence for an interaction between testosterone, immune function and carotenoid bioavailability in house sparrows and red-billed queleas: implications to testosterone handicap models of honest signaling. [Google Scholar]
- 15.During A., Dawson H. D., Harriosn E. H. 2005. Carotenoid transport is decreased and expression of the lipid transporters SR-BI, NPC1L1, and ABCA1 is downregulated in caco-2 Cells treated with ezetimibe. J. Nutr. 135, 2305–2312 [DOI] [PubMed] [Google Scholar]
- 16.Williams A. W., Boileau T. W. M., Erdman J. 1998. Factors influencing the uptake and absorption of carotenoids. Proc. Soc. Exp. Biol. Med. 218, 106–108 [DOI] [PubMed] [Google Scholar]
- 17.Eriksson J., et al. 2008. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010. 10.1371/journal.pgen.1000010 (doi:10.1371/journal.pgen.1000010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheesman D. F., Zagalsky P. F. 1966. Purification and properties of crustacyanin. Proc. R. Soc. Lond. B 164, 130–151 10.1098/rspb.1966.0017 (doi:10.1098/rspb.1966.0017) [DOI] [Google Scholar]
- 19.Kiefer C., Sumser E., Wernet M. F., von Lintig J. 2002. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl Acad. Sci. USA 99, 10 581–10 586 10.1073/pnas.162182899 (doi:10.1073/pnas.162182899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakudoh T., et al. 2010. A CD36-related transmembrane protein Is coordinated with an intracellular lipid-binding protein in selective carotenoid transport for cocoon coloration. J. Biol. Chem. 285, 7739–7751 10.1074/jbc.M109.074435 (doi:10.1074/jbc.M109.074435) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiefer C., Hessel S., Lampert J. M., Vogt K., Lederer M. O., Breithaupt D. E., von Lintig J. 2001. Identification and characterization of a mammalian enzyme catalyzing the asymmetric oxidative cleavage of provitamin A. J. Biol. Chem. 276, 14 110–14 116 10.1074/jbc.M011510200 (doi:10.1074/jbc.M011510200) [DOI] [PubMed] [Google Scholar]
- 22.Von Lintig J., Vogt K. 2000. Filling the gap in vitamin A research: molecular identification of an enzyme cleaving β-carotene to retinal. J. Biol. Chem. 275, 11 915–11 920 10.1074/jbc.275.16.11915 (doi:10.1074/jbc.275.16.11915) [DOI] [PubMed] [Google Scholar]
- 23.Wyss A., Wirtz G., Woggon W.-D., Brugger R., Wyss M., Friedlein A., Bachmann H., Hunziker W. 2000. Cloning and expression of β,β-carotene 15, 15′-dioxygenase. Biochem. Biophys. Res. Commun. 271, 334–336 10.1006/bbrc.2000.2619 (doi:10.1006/bbrc.2000.2619) [DOI] [PubMed] [Google Scholar]
- 24.During A., Doraiswamy S., Harrison E. H. 2008. Xanthophylls are preferentially taken up compared with β-carotene by retinal cells via a SRBI-dependent mechanism. J. Lipid Res. 49, 1715–1724 10.1194/jlr.M700580-JLR200 (doi:10.1194/jlr.M700580-JLR200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krieger M. 1999. Charting the fate of the ‘good cholesterol’: identification and characterization of the high-density lipoprotein receptor SR-BI. Annu. Rev. Biochem. 68, 523–558 10.1146/annurev.biochem.68.1.523 (doi:10.1146/annurev.biochem.68.1.523) [DOI] [PubMed] [Google Scholar]
- 26.Soccio R. E., Breslow J. L. 2003. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J. Biol. Chem. 278, 22 183–22 186 10.1074/jbc.R300003200 (doi:10.1074/jbc.R300003200) [DOI] [PubMed] [Google Scholar]
- 27.Tabunoki H., et al. 2004. A carotenoid-binding protein (CBP) plays a crucial role in cocoon pigmentation of silkworm (Bombyx mori) larvae. Insect Biochem. Mol. Biol. 28, 927–934 10.1016/j.febslet.2004.04.067 (doi:10.1016/j.febslet.2004.04.067) [DOI] [PubMed] [Google Scholar]
- 28.Watari H., Arakane F., Moog-Lutz C., Kallen C. B., Tomasetto C., Gerton G. L., Rio M. C., Baker M. E., Strauss J. F., III 1997. MLN64 contains a domain with homology to the steroidogenic acute regulatory protein (StAR) that stimulates steroidogenesis. Proc. Natl Acad. Sci. USA 94, 8462–8467 10.1073/pnas.94.16.8462 (doi:10.1073/pnas.94.16.8462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakudoh T., Tsuchida K., Kataoka H. 2005. BmStart1, a novel carotenoid-binding protein isoform from Bombyx mori, is orthologous to MLN64, a mammalian cholesterol transporter. Biochem. Biophys. Res. Commun. 336, 1125–1135 10.1016/j.bbrc.2005.08.241 (doi:10.1016/j.bbrc.2005.08.241) [DOI] [PubMed] [Google Scholar]
- 30.Bhosale P., Li B., Sharifzadeh M., Gellermann W., Frederick J. M., Tsuchida K., Bernstein P. S. 2009. Purification and partial characterization of a lutein-binding protein from human retina. Biochemistry 48, 4798–4807 10.1021/bi9004478 (doi:10.1021/bi9004478) [DOI] [PubMed] [Google Scholar]
- 31.Soccio R. E., Adams R. M., Romanowski M. J., Sehayek E., Burley S. K., Breslow J. L. 2002. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc. Natl Acad. Sci. USA. 99, 6943–6948 10.1073/pnas.052143799 (doi:10.1073/pnas.052143799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ganfornina M. D., Sanchez D., Pagano A., Tonachini L., Descalzi-Cancedda F., Martinez S. 2005. Molecular characterization and developmental expression pattern of the chicken apolipoprotein D gene: implications for the evolution of vertebrate lipocalins. Dev. Dyn. 232, 191–199 10.1002/dvdy.20193 (doi:10.1002/dvdy.20193) [DOI] [PubMed] [Google Scholar]
- 33.Wade N. M., Tollenaere A., Hall M. R., Degnan B. M. 2009. Evolution of a novel carotenoid-binding protein responsible for crustacean shell color. Mol. Biol. Evol. 26, 1851–1864 10.1093/molbev/msp092 (doi:10.1093/molbev/msp092) [DOI] [PubMed] [Google Scholar]
- 34.Londos C., Gruia-Gray J., Brasaemle D. L., Rondinone C. M., Takeda T., Dwyer N. K., Barber T., Kimmel A. R., Blanchette-Mackie E. J. 1996. Perilipin: possible roles in structure and metabolism of intracellular neutral lipids in adipocytes and steroidogenic cells. Int. J. Obes. Relat. Metab. Disord. 20, S97–S101 [PubMed] [Google Scholar]
- 35.Menon G. K., Menon J. 2000. Avian epidermal lipids: functional considerations and relationship to feathering. Am. Zool. 40, 540–552 10.1093/icb/40.4.540 (doi:10.1093/icb/40.4.540) [DOI] [Google Scholar]
- 36.Bhosale P., Larson A. J., Frederick J. M., Southwick K., Thulin C. D., Bernstein P. S. 2004. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J. Biol. Chem. 279, 49 447–49 454 10.1074/jbc.M405334200 (doi:10.1074/jbc.M405334200) [DOI] [PubMed] [Google Scholar]
- 37.Listowsky I., Abramovitz M., Homma H., Niitsu Y. 1988. Intracellular binding and transport of hormones and xenobiotics by glutathione-S-transferases. Drug Metab. Rev. 19, 305–318 10.3109/03602538808994138 (doi:10.3109/03602538808994138) [DOI] [PubMed] [Google Scholar]
- 38.Pemble S. E., Taylor J. B. 1992. An evolutionary perspective on glutathione transferases inferred from class-Theta glutathione transferase cDNA sequences. Biochem. J. 287, 957–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGraw K. J., Hudon J., Hill G. E., Parker R. S. 2005. A simple and inexpensive chemical test for behavioral ecologists to determine the presence of carotenoid pigments in animal tissues. Behav. Ecol. Sociobiol. 57, 391–397 10.1007/s00265-004-0853-y (doi:10.1007/s00265-004-0853-y) [DOI] [Google Scholar]
- 40.McGraw K. J., Adkins-Regan E., Parker R. S. 2002. Anhydrolutein in the zebra finch: a new, metabolically derived carotenoid in birds. Comp. Biochem. Physiol. B 132, 811–818 10.1016/S1096-4959(02)00100-8 (doi:10.1016/S1096-4959(02)00100-8) [DOI] [PubMed] [Google Scholar]
- 41.McGraw K. J. 2004. Colorful songbirds metabolize carotenoids at the integument. J. Avian Biol. 35, 471–476 10.1111/j.0908-8857.2004.03405.x (doi:10.1111/j.0908-8857.2004.03405.x) [DOI] [Google Scholar]
- 42.McGraw K. J., Hill G. E., Stradi R., Parker R. S. 2001. The influence of carotenoid acquisition and utilization on the maintenance of species-typical plumage pigmentation in male American goldfinches (Carduelis tristis) and northern cardinals (Cardinalis cardinalis). Physiol. Biochem. Zool. 74, 843–852 10.1086/323797 (doi:10.1086/323797) [DOI] [PubMed] [Google Scholar]
- 43.del Val E., Carlos J. C., Garrido-Fernandez J., Manuel J., Antoni B., Josep C., Negro J. J. 2009. Hepatic conversion of red carotenoids in passerine birds. Naturwissenschaften 96, 989–991 10.1007/s00114-009-0554-5 (doi:10.1007/s00114-009-0554-5) [DOI] [PubMed] [Google Scholar]
- 44.Kiriakidou M., McAllister J. M., Sugawara T., Strauss J. F., III 1996. Expression of steroidogenic acute regulatory protein (StAR) in the human ovary. J. Clin. Endocrinol. Metab. 81, 4122–4128.(doi:10.1210/jc.81.11.4122) [DOI] [PubMed] [Google Scholar]
- 45.Våge D. I., Boman I. A. 2010. A nonsense mutation in the beta-carotene oxygenase 2 (BCO2) gene is tightly associated with accumulation of carotenoids in adipose tissue in sheep (Ovis aries). BMC Genet. 11, 10(doi:10.1186/1471-2156-11-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berry S. D., et al. 2009. Mutation in bovine beta-carotene oxygenase 2 affects milk color. Genetics 182, 923–926 10.1534/genetics.109.101741 (doi:10.1534/genetics.109.101741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andersson S., Prager M., Johansson E. I. A. 2007. Carotenoid content and reflectance of yellow and red nuptial plumages in widowbirds (Euplectes spp.). Funct. Ecol. 21, 272–281 10.1111/j.1365-2435.2007.01233.x (doi:10.1111/j.1365-2435.2007.01233.x) [DOI] [Google Scholar]
- 48.Inouye C. Y., Hill G. E., Stradi R. D., Montgomerie R. 2001. Carotenoid pigments in male house finch plumage in relation to age, subspecies, and ornamental coloration. Auk 118, 900–915 10.1642/0004-8038(2001 (doi:10.1642/0004-8038(2001) [DOI] [Google Scholar]
- 49.McGraw K. J., Gregory A. J. 2004. Carotenoid pigments in male American goldfinches: what is the optimal biochemical strategy for becoming colourful? Biol. J. Linn. Soc. 83, 273–280 10.1111/j.1095-8312.2004.00388.x (doi:10.1111/j.1095-8312.2004.00388.x) [DOI] [Google Scholar]
- 50.Van der Horst D. J., Roosendaal S. D., Rodenburg K. W. 2009. Circulatory lipid transport: lipoprotein assembly and function from an evolutionary perspective. Mol. Cell. Biochem. 326, 105–119 10.1007/s11010-008-0011-3 (doi:10.1007/s11010-008-0011-3) [DOI] [PubMed] [Google Scholar]
- 51.Sugumaran M. 2002. Comparative biochemistry of eumelanogenesis and the protective roles of phenoloxidase and melanin in insects. Pigment Cell Res. 15, 2–9 10.1034/j.1600-0749.2002.00056.x (doi:10.1034/j.1600-0749.2002.00056.x) [DOI] [PubMed] [Google Scholar]
- 52.Connelly M. A. 2009. SR-BI-mediated HDL cholesteryl ester delivery in the adrenal gland. Mol. Cell. Biochem. 300, 83–88 10.1016/j.mce.2008.09.011 (doi:10.1016/j.mce.2008.09.011) [DOI] [PubMed] [Google Scholar]
- 53.Miller W. L. 2007. Steroidogenic acute regulatory protein (StAR), a novel mitochondrial cholesterol transporter. Biochim. Biophys. Acta 1771, 663–676 [DOI] [PubMed] [Google Scholar]
- 54.Olson J. A. 1984. Serum levels of vitamin A and carotenoids as reflectors of nutritional status. J. Natl Cancer I 73, 1439–1444 [PubMed] [Google Scholar]
- 55.Tajima S., Goda T., Takase S. 2001. Co-ordinated induction of β-carotene cleavage enzyme and retinal reductase in the duodenum of the developing chicks. Comp. Biochem. Phys. B 128, 425–434 10.1016/S1096-4959(00)00347-X (doi:10.1016/S1096-4959(00)00347-X) [DOI] [PubMed] [Google Scholar]
- 56.Reboul E., et al. 2005. Lutein transport by Caco-2 TC-7 cells occurs partly by a facilitated process involving the scavenger receptor class B type I (SR-BI). Biochem. J. 387, 455–461 10.1042/BJ20040554 (doi:10.1042/BJ20040554) [DOI] [PMC free article] [PubMed] [Google Scholar]