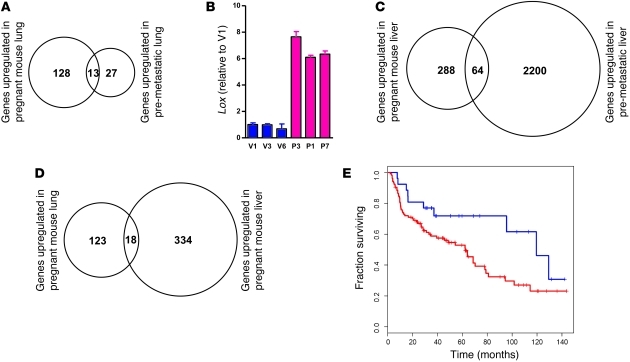
Figure 8. Gene expression profiling of pregnant versus virgin mouse lung and comparative dataset analysis reveal similarities with a premetastatic lung signature characterized by accumulation of myelomonocytic cells.
RNA from 2 mice each (16-day pregnant and virgin NOD/SCID) were pooled for 1 Affymetrix mouse 430 chip, and 3 chips were used per condition. Differentially expressed genes in the lungs and livers were determined by the rank product method by setting the false detection rate (FDR) to 5%. (A) Comparative analysis of upregulated genes in pregnant mouse lung with upregulated genes in premetastatic lung (mice harboring aggressive versus benign primary tumors; data derived from the heatmap of Supplemental Figure 1D in ref. 10). Overlap P = 4.1 × 10–20. (B) Quantitative RT-PCR for Lox in lung RNA of 16-day pregnant (P-) and virgin (V-) NOD/SCID mice. (C) Comparative analysis of upregulated genes in pregnant mouse liver with upregulated genes (>2 fold change) in premetastatic (untreated) liver (data derived from ref. 28). Overlap P = 1.3 × 10–6. (D) Commonly upregulated genes in pregnant mouse lung and liver. Overlap P = 1.1 × 10–11. (E) Unsupervised hierarchical clustering of lung cancer patients (from ref. 29) using orthologs of downregulated genes in pregnant mouse lung followed by Kaplan-Meier analysis of the 2 main patient groups (red, poor prognosis; blue, good prognosis). P = 0.0423. See also Supplemental Table 2.