Figure 3.
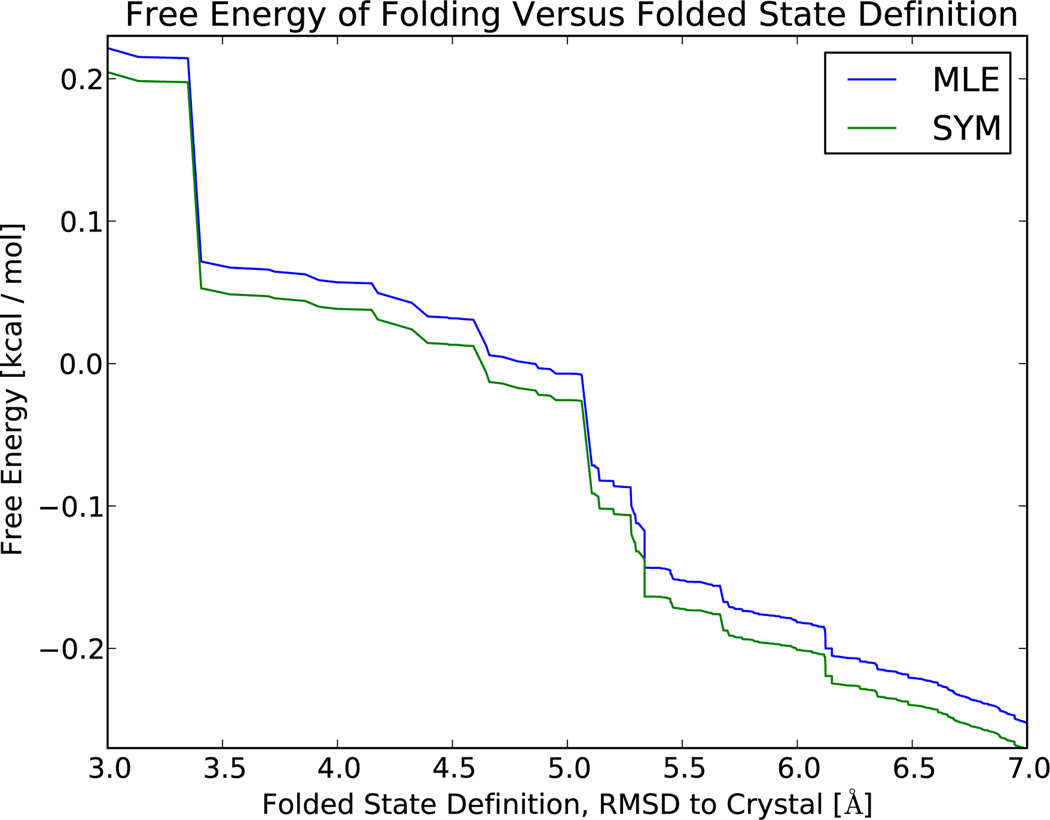
Simulations of the WW protein12 were used to compare the performance of the symmetrized and MLE protocols. Folding free energies calculated using a two-state approximation , show good agreement (Δ ≤ 0.03 kcal / mol) between models constructed using the symmetrized and MLE protocols, as expected for long trajectories. The near-zero folding free energy is expected, as the simulations were performed near the melting temperature;12 the exact free energy depends weakly on how one defines the folded state. Here, the folded state is defined as all states with an RMSD (to crystal structure) below some cutoff value; the unfolded state is defined as the remaining states. The large RMSD values observed are due to the large conformational fluctuations observed in the high temperature (393 K) simulations.