Table 5.
A summary of the missing values for each of the tests and the number of significant genes detected by other methods within those missing values.
| Significant Genes from Sets of Missing Genes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Method | EBA (Affymetrix) |  (LRT) (LRT) |
EBA (RNA-Seq) | edgeR | DESeq |
|
|
|
| EBA (Affymetrix) | 14292 | 561 | 17 | 589 | 450 | 16 | 2550 | 1316 |
 (LRT) (LRT) |
75 | 3979 | 0 | 235 | 197 | 0 | 3064 | 2208 |
| EBA (RNA-Seq) | 81 | 0 | 4726 | 235 | 197 | 0 | 3064 | 2208 |
| edgeR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| DESeq | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
 |
422 | 973 | 5 | 1166 | 997 | 8947 | 4171 | 2406 |
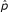 |
2 | 0 | 0 | 0 | 0 | 0 | 915 | 0 |
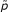 |
3 | 0 | 0 | 0 | 0 | 0 | 856 | 1832 |
The diagonal gives the number of genes with missing tests. The off-diagonals indicate those genes that are significant for one method amongst the missing calls for another method. The acronym LRT denotes the likelihood ratio test based on the Poisson distribution, as described in the Methods. The acronym EBA denotes the empirical Bayes analysis performed on both the Affymetrix and RNA-Seq data.
