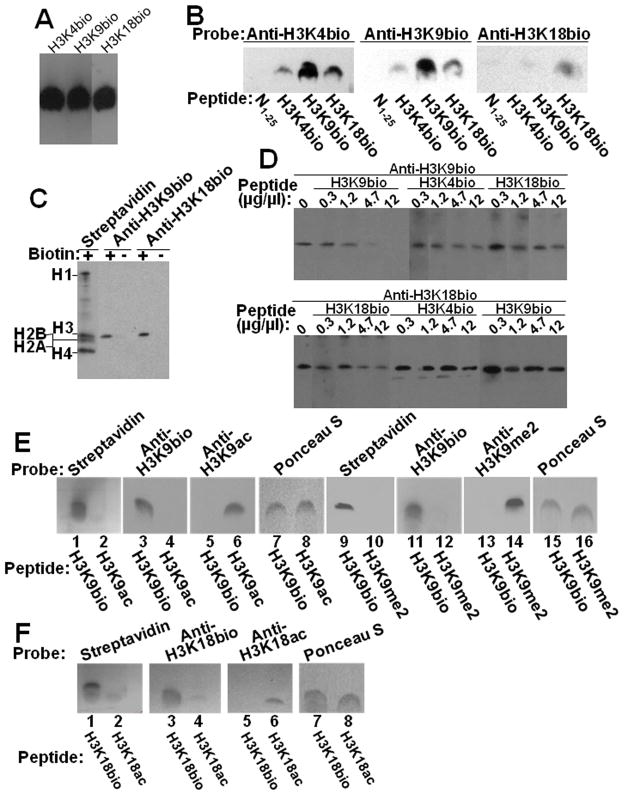
Fig. 3. Specificity of antibodies to H3K4bio, H3K9bio and H3K18bio.
Panel A: Confirmation of equal loading of peptides H3K4bio, H3K9bio, and H3K18bio with streptavidin. Panel B: Transblots of peptides N1–25 (non-biotinylated negative control), H3K4bio, H3K9bio, and H3K18bio were probed with anti-H3K4bio (left), anti-H3K9bio (middle), and anti-H3K18bio (right). Panel C: HCl extracts of histones from Jurkat cells were probed using streptavidin, anti-H3K9bio, and anti-H3K18bio; samples of biotin-free histones (“-”) were generated by using avidin agarose. Panel D: Bulk HCl extracts of histones from Jurkat cells were probed with anti-H3K9bio (top) and anti-H3K18bio (bottom) after pre-incubation of antibodies with increasing amounts of competing peptides H3K4bio, H3K9bio, and H3K18bio; controls (“C”) were prepared without peptide competitors. Note the difference in the order of peptide competitors in the two gels. For some gels, bands from the same analytical runs were electronically re-arranged to facilitate comparisons. Panel E: Peptides H3K9bio, H3K9ac, and H3K9me2 were probed with streptavidin (lanes 1, 2, 9 and 10), anti-H3K9bio (lanes 3, 4, 11 and 12), anti-H3K9ac (lanes 5 and 6), and anti-H3K9me2 (lanes 13 and 14); Ponceau S was used as loading control (lanes 7, 8, 15 and 16). Panel F: Peptides H3K18bio and H3K18ac were probed with streptavidin (lanes 1 and 2), anti-H3K18bio (lanes 3 and 4) and anti-H3K18ac (lanes 5 and 6); Ponceau S was used as loading control (lanes 7 and 8).