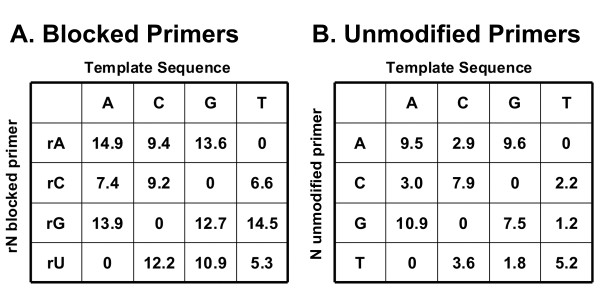
Figure 7.
Mismatch discrimination using blocked-cleavable primers with an RNA:DNA base-pair mismatch versus standard unmodified allele-specific PCR primers. Four synthetic 103 base oligonucleotide targets were employed where a single base was varied (A, C, G, or T) within the primer binding site. For each DNA target, a single common For primer was paired with four different "rDDDDx" blocked-cleavable Rev primers, varying the RNA base (A), or with four unmodified allele-specific primers terminating in a different 3'-DNA base (B). Amplification reactions were performed in real-time mode using SYBR® Green detection; all reactions were run in triplicate. ΔCq values are shown in the tables and represent the difference between the Cq value for the allele-specific primer for each template and the Cq value for the perfect match template. The concentration of RNase H2 was 1.3 mU per 10 μL.