Abstract
The stickleback family (Gasterosteidae) of fish is less than 40 million years old, yet stickleback species have diverged in both diploid chromosome number (2n) and morphology. We used comparative fluorescence in situ hybridization (FISH) on 2 stickleback species, Gasterosteus aculeatus (2n = 42) and Apeltes quadracus (2n = 46), to ascertain the types of chromosome rearrangements that differentiate these species. The A. quadracus karyotype contains more acrocentric and telocentric chromosomes than the G. aculeatus karyotype. By using bacterial artificial chromosome probes from G. aculeatus in our FISH screen, we found that 6 pericentric inversions and 2 chromosome fusions/fissions are responsible for the greater number of acrocentric and telocentric chromosomes in A. quadracus. While most populations of G. aculeatus have an XX/XY sex chromosome system, A. quadracus has a ZZ/ZW sex chromosome system, as previously reported. However, we discovered that a population of A. quadracus from Connecticut lacks heteromorphic sex chromosomes, providing evidence for unexpected sex chromosome diversity in this species.
Key Words: Chromosome evolution, Fish sex chromosome, Fluorescence in situ hybridization, Karyotype differentiation, Stickleback
Introduction
Stickleback fishes are small teleosts of the family Gasterosteidae, which comprises 6 species that inhabit temperate freshwater and marine environments across the northern hemisphere [Wootton, 1976]. Previous studies on chromosome number and morphology revealed diverse karyotypes among stickleback species [Chen and Reisman, 1970; Ocalewicz et al., 2011] as well as within a single species [Kitano et al., 2009]. Stickleback species also have different sex chromosome systems. Both threespine sticklebacks (Gasterosteus aculeatus) and ninespine sticklebacks (Pungitius pungitius) have XX/XY (male heteromorphic) sex chromosome systems, but comparative fluorescence in situ hybridization (FISH) mapping and genetic linkage analyses indicate that these sex chromosome systems arose independently [Peichel et al., 2004; Ocalewicz et al., 2008; Ross and Peichel, 2008; Ross et al., 2009; Shapiro et al., 2009]. A third species, the fourspine stickleback (Apeltes quadracus), has a ZZ/ZW (female heteromorphic) sex chromosome system [Chen and Reisman, 1970; Ross et al., 2009]. In addition, the black-spotted stickleback (G. wheatlandi) and G. aculeatus populations from the Sea of Japan each have independently evolved X1X1X2X2/X1X2Y sex chromosome systems [Kitano et al., 2009; Ross et al., 2009].
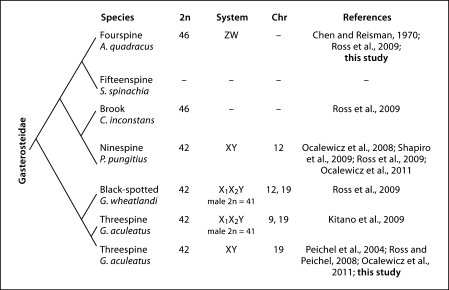
The phylogenetic relationships between species in the stickleback family have previously been disputed [Mattern, 2004]. However, a recent comprehensive phylogeny has resolved the evolutionary relationship among the sticklebacks [Kawahara et al., 2009]. When all available karyotype data for sticklebacks are mapped onto this phylogeny, it appears that sticklebacks have diverged more rapidly in karyotype than previously appreciated, since even closely related stickleback species have different diploid chromosome numbers, karyotypes, and sex chromosome systems (fig. 1).
Fig. 1.
A phylogeny of the family Gasterosteidae, incorporating major findings from this study and other reports. The phylogeny is based on Kawahara et al. [2009], though branch lengths are arbitrary. Listed are the common name and scientific name for each stickleback species, as well as the diploid number, the heteromorphic sex chromosome system (if known), and the chromosome on which the sex determination gene is found (if known). Chromosomes in all stickleback species are numbered in reference to G. aculeatus.
To explore the rapid evolution of stickleback karyotypes in more detail, we designed a comprehensive FISH survey to identify the major genomic rearrangements that have occurred in the karyotypes of 2 stickleback species since they diverged from a common ancestor. We chose 2 species with different diploid chromosome numbers and divergent karyotypes, the threespine stickleback (G. aculeatus) and the fourspine stickleback (A. quadracus) [Chen and Reisman, 1970; Ross et al., 2009]. In addition, while G. aculeatus has an XX/XY sex chromosome system found on chromosome 19 [Peichel et al., 2004; Ross and Peichel, 2008], the ancestral chromosome pair that evolved into the A. quadracus ZZ/ZW sex chromosome pair is not yet known. Thus, we designed our survey to uncover the genomic rearrangements that generated the diverse karyotypes of these species and to reveal the identity of the A. quadracus ZZ/ZW sex chromosome pair.
Material and Methods
Fish Collection and Breeding
We collected male and female A. quadracus from the West River Memorial Park in New Haven, Connecticut, in May 2009. These males and females were brought into the laboratory and allowed to breed naturally in 110-liter aquarium tanks in July and August 2009. We grew progeny from these matings together in 474-liter aquarium tanks until adulthood. Tissue from both wild-caught A. quadracus individuals and their progeny was used to prepare metaphase chromosome spreads (see below). In April and June 2010, we collected male G. aculeatus from 2 locations in Lake Washington: Union Bay in Seattle, Washington and the Mercer Slough Nature Park in Bellevue, Washington (Washington permit 10-049). We housed these fish in 110-liter aquarium tanks. Metaphase chromosome spreads were prepared using tissue from wild-caught G. aculeatus individuals (see below). For both species, we kept fish in summer lighting conditions (16 h light, 8 h dark) at approximately 16°C in 0.35% saltwater (3.5 g/l Instant Ocean salt (Aquarium Systems, Mentor, Ohio, USA); 0.4 ml/l NaHCO3). We fed the fish with live brine shrimp nauplii twice daily. All procedures were approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1575).
Selection of FISH Probes
We used G. aculeatus bacterial artificial chromosome (BAC) clones from the CHORI-215 library [Kingsley et al., 2004] as probes for FISH (table 1). To identify BAC probes for each end of each G. aculeatus chromosome pair, we extracted 200 kb of genomic sequence from the end of each G. aculeatus chromosome assembly (BROAD S1 assembly, February 2006, available at http://www.ensembl.org/Gasterosteus_aculeatus/index.html) and used this sequence in a BLAST search of the publicly available BAC end sequences [Kingsley and Peichel, 2007]. If both the T7 and SP6 end sequence reads of the BAC clones aligned within the 200-kb genomic sequence we selected, we used those paired end sequence reads from that BAC in a BLAT search against the G. aculeatus genome. If the paired end sequence reads of the BAC clone aligned to the desired chromosome assembly in opposing orientation, with expected separation based on the average size of a CHORI-215 BAC insert [Kingsley et al., 2004], we then used the BAC clone as a FISH probe for that G. aculeatus chromosome (table 1).
Table 1.
BAC clones used as FISH probes in this study
| Chromosome | Length, bp | BAC | Color | Location, bp |
|---|---|---|---|---|
| 1 | 28,185,914 | CH215–01F04 | purple | 808,402–984,867 |
| CH215-36H04 | green | 26,520,970–26,685,556 | ||
| 2 | 23,295,652 | CH215-14A06 | purple | 1,256,893–1,362,297 |
| CH215-51B03 | green | 22,208,609–22,374,157 | ||
| 3 | 16,798,506 | CH215-22E01 | purple | 904,453–1,069,729 |
| CH215-18O23 | green | 14,811,263–14,979,917 | ||
| 4 | 32,632,948 | CH215-37J16 | purple | 1,324,641–1,484,145 |
| CH215-03K06 | green | 31,667,549–31,830,249 | ||
| 5 | 12,251,397 | CH215-10F03 | purple | 906,711–1,099,727 |
| CH215-15A11 | green | 11,086,429–11,245,223 | ||
| 6 | 17,083,675 | CH215-39G07 | purple | 1,009,878–1,175,833 |
| CH215-30J20 | green | 16,083,765–16,163,690 | ||
| 7 | 27,937,443 | CH215-24C20 | purple | 1,118,953–1,279,901 |
| CH215-50F19 | green | 26,759,364–26,929,269 | ||
| 8 | 19,368,704 | CH215-18K21 | purple | 1,728,015–1,897,447 |
| CH215-50P04 | green | 18,234,424–18,411,738 | ||
| 9 | 20,249,479 | CH215-09O14 | purple | 1,205,121–1,367,430 |
| CH215-35K20 | green | 18,760,858–18,914,793 | ||
| 10 | 15,657,440 | CH215-32N20 | purple | 939,436–1,124,528 |
| CH215-13K03 | green | 14,608,763–14,740,402 | ||
| 11 | 16,706,052 | CH215-22E24 | purple | 1,328,616–1,481,096 |
| CH215-32F20 | green | 15,594,645–15,722,431 | ||
| 12 | 18,401,067 | CH215-35E14 | purple | 951,837–1,050,566 |
| CH215-19P17 | green | 17,405,092–17,588,100 | ||
| 13 | 20,083,130 | CH215-29N20 | purple | 1,278,281–1,129,621 |
| CH215-41J19 | green | 18,303,573–18,452,590 | ||
| 14 | 15,246,461 | CH215-50I22 | purple | 1,523,325–1,641,391 |
| CH215-04P19 | green | 14,054,345–14,228,449 | ||
| 15 | 16,198,764 | CH215-42G09 | purple | 1,136,990–1,288,015 |
| CH215-47G02 | green | 14,910,768–15,091,535 | ||
| 16 | 18,115,788 | CH215-25D03 | purple | 5,417,553–5,595,117 |
| CH215-59O23 | green | 15,244,265–15,412,230 | ||
| 17 | 14,603,141 | CH215-03L14 | purple | 1,687,087–1,798,208 |
| CH215-25N10 | green | 13,556,132–13,679,389 | ||
| 18 | 16,282,716 | CH215-60K06 | purple | 2,420,846–2,594,158 |
| CH215-58L04 | green | 14,897,105–15,061,421 | ||
| 19 | 20,240,660 | CH215-23N18 | purple | 849,540–1,016,623 |
| CH215-16P13 | green | 4,756,773–4,923,988 | ||
| 20 | 19,732,071 | CH215-32D17 | purple | 16,730,161–16,890,598 |
| CH215-09B01 | green | 18,782,835–18,919,085 | ||
| 21 | 11,717,487 | CH215-11F14 | purple | 982,376–1,039,352 |
| CH215-34H24 | green | 10,642,915–10,791,824 | ||
For each G. aculeatus chromosome, the total length in the G. aculeatus genome assembly (version BROAD S1, February 2006, http://www.ensembl.org/Gasterosteus_aculeatus/index.html) is given. The pseudocolor for each BAC clone is indicated, as are the positions of the ends of each BAC clone in the respective chromosome assembly.
For 18 of 21 G. aculeatus chromosomes, the selected BAC clones aligned to a region within 2.6 Mb of the predicted ends of that chromosome assembly and hybridized to the ends of chromosomes in G. aculeatus male metaphase spreads. For 3 chromosomes, 1 BAC clone aligned to a genomic sequence within 2.6 Mb of the predicted ends of that chromosome assembly, but did not hybridize to the end of a chromosome pair in G. aculeatus male metaphase spreads. In those 3 cases, we screened additional 200-kb regions of those chromosome assemblies until we identified BAC clones that met the above selection criteria and hybridized to the ends of a chromosome pair in male G. aculeatus metaphase spreads. For the chromosomes involved (chromosomes 16, 19 and 20), errors in the G. aculeatus genome assembly, as previously reported for chromosome 19 [Ross and Peichel, 2008], or population-specific chromosome rearrangements could account for these observations.
Cytogenetic Analysis
We prepared metaphase spreads as described previously [Ross and Peichel, 2008] with the following modifications. We performed intraperitoneal injections of 5 μl (A. quadracus) or 10 μl (G. aculeatus) of 1% colchicine in phosphate-buffered saline into adult fish and incubated them for 12–14 h in an aquarium. After euthanizing the fish in 0.025% tricaine methanesulfonate (MS-222), we determined the sex of individual fish by examination of the gonads. We combined spleen tissue from several individuals to make metaphase spreads of males and females from both species (table 2).
Table 2.
Number of individual samples and pools analyzed by FISH in A. quadracus and G. aculeatus and karyotyped in A. quadracus
| FISH analyses |
Karyotypic analyses |
|||||
|---|---|---|---|---|---|---|
| total pools analyzed | individuals/pool (range) | metaphases analyzed/chromosome (range) | total pools analyzed | individuals/pool (range) | metaphases analyzed | |
| A. quadracus ♀ | 8 | 12 (9–17) | 5 (3–10) | 3 | 11 (7–15) | 6 |
| A. quadracus ♂ | 7 | 8 (7–11) | 3 (3–5) | 2 | 8 (6–11) | 5 |
| G. aculeatus ♂ | 5 | 21 (15–36) | 3 (3–4) | N/A | N/A | N/A |
N/A = Not applicable.
We conducted FISH as previously described [Ross and Peichel, 2008], with the following modifications. We extracted and purified BAC DNA using an AutoGen 740 automated system (AutoGen, Holliston, Mass., USA) and quantified BAC DNA by gel electrophoresis. Using the Vysis nick translation kit (Abbott Labs, North Chicago, Ill., USA), we labeled 1 μg of each BAC clone with either ChromaTide Alexa Fluor 488-5-dUTP or 568-5-dUTP (Invitrogen, Carlsbad, Calif., USA). Two hundred nanograms of the labeled clones were ethanol precipitated with 10 μg salmon sperm DNA and 10 μg genomic DNA from the species used in the experiment. We allowed hybridization to proceed for 1–2 nights at 37°C. We viewed washed slides using the 100× objective on a Nikon Eclipse 80i microscope (Nikon, Shinjuku, Tokyo, Japan) with an automated filter turret using Chroma filters 31000v2 (DAPI), 41001 (FITC), and 41004 (Texas Red) (Chroma, Bellows Falls, Vt., USA). We captured images using a Photometrics Coolsnap ES2 camera (Photometrics, Tucson, Ariz., USA) and used NIS Elements imaging software (BR 3.00, SP7, Hotfix8, Build 548, Nikon, Shinjuku, Tokyo, Japan) to pseudocolor those images grey (DAPI), green (Alexa 488) and purple (Alexa 568).
Chromosome Morphology
For G. aculeatus and A. quadracus, we used images from FISH experiments (figs. 2, 3; data not shown) to assess the morphology of each chromosome. We classified chromosomes according to this average arm length ratio (long arm:short arm, L:S). Chromosomes were classified as metacentric (1.0–1.7), submetacentric (1.7–3.0), acrocentric (3.0–7.0), or telocentric (7.0 or greater), following Levan et al. [1964] and Schrader et al. [1997] (table 3). The only exception to this method is G. aculeatus chromosome 19, which is a heteromorphic XX/XY sex chromosome pair [Peichel et al., 2004; Ross and Peichel, 2008]. Thus, we measured the arm lengths of the single X chromosome in male G. aculeatus, but did not measure the Y chromosome (fig. 3; data not shown). For all FISH experiments and karyotype analyses, we examined multiple metaphase spreads from multiple pools of individual fish (table 2). Based on the chromosome morphology, relative centromere position, and BAC probe hybridization positions for each chromosome in our FISH screen, we determined the types of chromosome rearrangements that could explain the observed differences between the G. aculeatus and A. quadracus karyotypes (table 3).
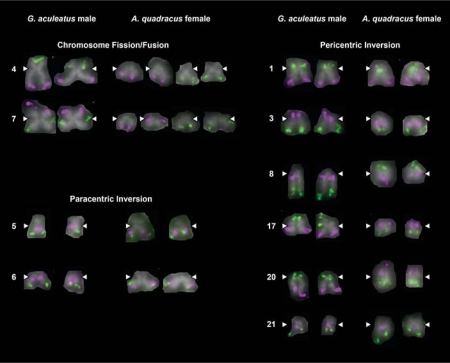
Fig. 2.
Major chromosomal rearrangements between G. aculeatus and A. quadracus. Two-color FISH images are shown for the 10 G. aculeatus chromosomes that show visible evidence for chromosome rearrangement. Pairs of homologs from individual metaphase spreads of G. aculeatus males and A. quadracus females are shown. White arrowheads indicate the position of the centromere.
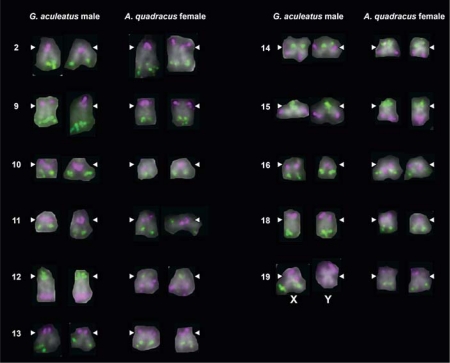
Fig. 3.
No major differences in chromosome morphology or probe hybridization between 11 G. aculeatus and A. quadracus chromosome pairs. Two-color FISH images are shown for both homologs of each chromosome pair. White arrowheads indicate the position of the centromere. One FISH probe for chromosome 19 does not hybridize to the metacentric Y chromosome, most likely due to a large deletion on the Y chromosome in G. aculeatus [Ross and Peichel, 2008].
Table 3.
Chromosome morphology and evidence for chromosome rearrangements differentiating the chromosomes of G. aculeatus and A. quadracus
| G. aculeatus chromosome | Mean arm ratio (L:S) | Chromosome morphology | A. quadracus chromosome | Mean arm ratio (L:S) | Chromosome morphology | Proposed rearrangement |
|---|---|---|---|---|---|---|
| 1 | 1.90 | submetacentric | 1 | 3.33 | acrocentric | pericentric inversion |
| 2 | 5.28 | acrocentric | 2 | 4.30 | acrocentric | |
| 3 | 4.67 | acrocentric | 3 | 5.10 | acrocentric | pericentric inversion |
| 4 | 2.15 | submetacentric | 4a | 3.39 | acrocentric | fission/fusion |
| 4b | 5.04 | acrocentric | ||||
| 5 | 2.13 | submetacentric | 5 | 1.98 | submetacentric | paracentric inversion |
| 6 | 4.38 | acrocentric | 6 | 5.92 | acrocentric | paracentric inversion |
| 7 | 1.23 | metacentric | 7a | 4.88 | acrocentric | fission/fusion |
| 7b | 6.52 | acrocentric | ||||
| 8 | 9.17 | telocentric | 8 | 1.32 | metacentric | pericentric inversion |
| 9 | 6.04 | acrocentric | 9 | 3.41 | acrocentric | |
| 10 | 13.68 | telocentric | 10 | 3.48 | acrocentric | |
| 11 | 5.37 | acrocentric | 11 | 11.65 | telocentric | |
| 12 | 5.51 | acrocentric | 12 | 3.77 | acrocentric | |
| 13 | 5.74 | acrocentric | 13 | 3.08 | acrocentric | |
| 14 | 1.49 | metacentric | 14 | 1.36 | metacentric | |
| 15 | 9.97 | telocentric | 15 | 3.06 | acrocentric | |
| 16 | 9.73 | telocentric | 16 | 8.12 | telocentric | |
| 17 | 1.42 | metacentric | 17 | 3.23 | acrocentric | pericentric inversion |
| 18 | 6.19 | acrocentric | 18 | 3.52 | acrocentric | |
| 19 (X) | 2.67 | submetacentric | 19 | 2.44 | submetacentric | |
| 20 | 9.88 | telocentric | 20 | 1.36 | metacentric | pericentric inversion |
| 21 | 1.39 | metacentric | 21 | 3.36 | acrocentric | pericentric inversion |
We also calculated the fundamental number (major chromosome arm number; NF) for both sexes of G. aculeatus and A. quadracus [Matthey, 1949; Klinkhardt, 1998]. For calculating NF, we followed White [1978] and Klinkhardt [1998] and considered that metacentric and submetacentric chromosomes each have 2 major chromosome arms, while acrocentric and telocentric chromosomes each have 1 major chromosome arm. We inferred NF and other karyotype features for G. aculeatus females (table 4) based on the absence of the metacentric Y and the presence of a second submetacentric X chromosome [Ross and Peichel, 2008].
Table 4.
Major features of the karyotypes of G. aculeatus and A. quadracus
| Species | Sex | 2n | Chromosomes |
NF | |||
|---|---|---|---|---|---|---|---|
| metacentric | submetacentric | acrocentric | telocentric | ||||
| G. aculeatus | female | 42 | 8 | 6+2X | 16 | 10 | 58 |
| male | 42 | 8+Y | 6+X | 16 | 10 | 58 | |
| A. quadracus | female | 46 | 6 | 4 | 32 | 4 | 56 |
| male | 46 | 6 | 4 | 32 | 4 | 56 | |
2n = Diploid number; NF = fundamental number.
Results
FISH Screen Identifies the Chromosome Morphology of Each G. aculeatus Chromosome Pair
The G. aculeatus male karyotype consists of 42 chromosomes: 8 autosomes are metacentric, 6 are submetacentric, 16 are acrocentric, 10 are telocentric, and there is 1 submetacentric X and 1 metacentric Y (table 4) [Ross and Peichel, 2008]. The relatively high NF (58) for G. aculeatus compared to its diploid number (2n = 42) reflects a relatively large number of metacentric and submetacentric chromosomes in the karyotype. The selected BAC clones (table 1) hybridize by FISH to the ends of each chromosome (figs. 2, 3). By combining FISH with measurements of chromosome arm length, we were able to assign each G. aculeatus chromosome pair to a morphological class (table 3).
FISH Screen Identifies the Major Differences between the G. aculeatus and A. quadracus Karyotypes
We used the same BAC clones selected for the G. aculeatus male FISH analysis in a similar survey of the chromosomes in A. quadracus females and males (figs. 2, 3). The A. quadracus karyotype consists of 46 chromosomes, of which 6 chromosomes are metacentric, 4 are submetacentric, 32 are acrocentric, and 4 are telocentric (fig. 4; table 4). The dominance of acrocentric and telocentric chromosomes in the A. quadracus karyotype (78%) is reflected in a lower ratio of NF to diploid chromosome number; NF:2n is 56:46 for A. quadracus, while it is 58:42 for G. aculeatus (table 4).
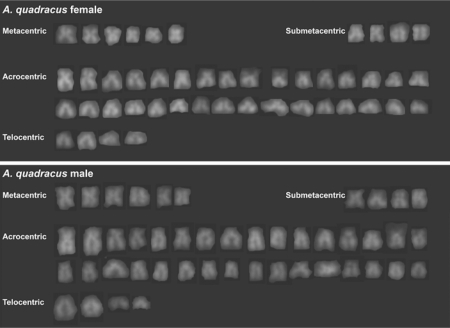
Fig. 4.
DAPI-stained karyotypes of an A. quadracus female (upper panel) and an A. quadracus male (lower panel) from the West River in New Haven, Connecticut.
Two of the larger chromosome pairs in the G. aculeatus karyotype, chromosomes 4 (submetacentric) and 7 (metacentric), each correspond to 2 pairs of relatively small acrocentric chromosomes in A. quadracus (fig. 2; table 3). Eight chromosome pairs appear to have undergone inversions in the time since G. aculeatus and A. quadracus diverged; 6 are pericentric inversions, which encompass the centromere, and 2 are paracentric inversions, which do not encompass the centromere (fig. 2; table 3). Five of these pericentric inversions have resulted in visibly different morphology for the orthologous G. aculeatus and A. quadracus chromosomes. Probes from 11 G. aculeatus chromosome pairs show identical hybridization patterns and morphologies in both G. aculeatus and A. quadracus (fig. 3).
A. quadracus Males and Females from Connecticut Do Not Have Heteromorphic Sex Chromosomes
A heteromorphic ZZ/ZW sex chromosome pair has been described in A. quadracus populations from Maine [Chen and Reisman, 1970] and Massachusetts [Ross et al., 2009]. However, we found no evidence for a heteromorphic sex chromosome pair in A. quadracus male or female progeny from specimens collected in the West River in New Haven, Connecticut (fig. 4). In addition, hybridization patterns were identical in metaphase spreads from male and female A. quadracus for all FISH probes used in this study (data not shown).
Discussion
Diversity of the A. quadracus Sex Chromosomes
We undertook a comparative FISH study to identify the major genomic rearrangements that have occurred in lineages separating 2 stickleback species, A. quadracus and G. aculeatus. These species are intriguing subjects for such a study, not only because of reported differences in karyotype, but also due to the divergence of sex chromosome systems [Chen and Reisman, 1970; Ross et al., 2009]. In G. aculeatus, chromosomes 19 are an XX/XY sex chromosome pair [Peichel et al., 2004; Ross and Peichel, 2008]. Two previous studies have shown that A. quadracus populations from Maine and Massachusetts have a ZZ/ZW sex chromosome pair [Chen and Reisman, 1970; Ross et al., 2009]. In both studies, the W chromosome is described as a large acrocentric chromosome, while the Z chromosome is also acrocentric, but has a smaller long arm than the W chromosome. Genetic markers from chromosome 19 are not linked to the sex determination locus in an A. quadracus family from Massachusetts [Ross et al., 2009], and no genetic markers linked to a sex determination locus have been found in A. quadracus.
In the present study of A. quadracus from Connecticut, we find that neither females nor males have a heteromorphic chromosome pair indicative of either a ZZ/ZW or XX/XY sex chromosome system. Two possible explanations could account for the differences in sex chromosomes between the Connecticut population of A. quadracus and the Maine and Massachusetts populations. First, A. quadracus from Connecticut could have a cryptic (homomorphic) ZZ/ZW sex chromosome pair that is homologous to the ZZ/ZW pair in the Massachusetts and Maine populations. The larger size of the W chromosome relative to the Z chromosome in those populations [Chen and Reisman, 1970; Ross et al., 2009] could be due to an increase in the amount of heterochromatin in the W chromosome relative to the Z chromosome following the cessation of recombination with the Z chromosome. If negligible amounts of heterochromatin were found in the A. quadracus W chromosome of the Connecticut population, then the Z and W chromosomes in this population would not be morphologically distinguishable. Such a population-specific difference in the amount of heterochromatin or repetitive DNA sequences present on the sex chromosomes has been reported in the guppy Poecilia reticulata [Hornaday et al., 1994], the platyfish Xiphophorus maculatus [Nanda et al., 2000], and the glass knifefish Eigenmannia virescens [Almeida-Toledo et al., 2001]. Differences in the amount of heterochromatin on the sex chromosomes have also been reported between closely related species of salmonids [Moran et al., 1996] and South American catfishes [Andreata et al., 1992].
Second, A. quadracus from Connecticut could have a different sex determination mechanism than the Massachusetts and Maine ZZ/ZW sex chromosome system. This mechanism could be genetic, environmental, or a combination of genetic and environmental signals. Extreme variation in the mechanism of sex determination within a single species is not without precedent. In the Atlantic silverside, Menidia menidia, both environmental and genetic sex determination mechanisms are present and vary by latitude [Conover and Heins, 1987]. Although we currently have little data on the ecology or population structure of A. quadracus, it will be interesting to determine whether variation in sex determination mechanisms is associated with any ecological or geographical factors in this species.
Even though the A. quadracus population in this study does not have a heteromorphic ZZ/ZW sex chromosome system, our comparative FISH study could help to identify the ZZ/ZW sex chromosome system in A. quadracus. The ZZ/ZW pair in both Massachusetts and Maine populations of A. quadracus is one of the larger chromosome pairs in the karyotype [Chen and Reisman, 1970; Ross et al., 2009]. In addition, the karyotypes of the Connecticut and Massachusetts A. quadracus are similar, differing only in the presence of the large W chromosome in A. quadracus females from the Massachusetts population [Ross et al., 2009]. Thus, the heteromorphic ZZ/ZW pair in the Massachusetts A. quadracus karyotype could correspond to one of the larger chromosome pairs in the Connecticut A. quadracus karyotype. Our FISH experiments showed that chromosomes 1, 2, 9, 12, 13, 19, and 20 were among the larger chromosome pairs in their respective metaphase spreads. Two of these (chromosomes 12 and 19) have previously been ruled out as the Massachusetts A. quadracus ZZ/ZW pair [Ross et al., 2009]; however, the other large chromosomes remain targets for further studies to identify the ZZ/ZW sex chromosome pair in A. quadracus.
Population differences in sex chromosomes and sex determination mechanisms are also found in other species of fish. Studies have documented sex chromosome diversity within species of poeciliid fish [Nanda et al., 2000; Volff and Schartl, 2001; Schultheis et al., 2009] and within several species of neotropical fish, including E. virescens [Almeida-Toledo et al., 2002; Henning et al., 2011], Erythrinus erythrinus [Bertollo et al., 2004; Cioffi et al., 2010], and Hoplias malabaricus [Bertollo et al., 2000; Cioffi and Bertollo, 2010]. Sex chromosome systems also differ among populations of other stickleback species. G. aculeatus from the Sea of Japan have an X1X1X2X2/X1X2Y sex chromosome system, which likely arose from the ancestral XX/XY sex chromosome system found in other G. aculeatus populations [Kitano et al., 2009]. Black-spotted sticklebacks (G. wheatlandi) from a population in Maine have an XX/XY sex chromosome system [Chen and Reisman, 1970], while at least 1 G. wheatlandi population in Massachusetts has an X1X1X2X2/X1X2Y sex chromosome system [Ross et al., 2009]. Future studies should reveal whether most A. quadracus populations have ZZ/ZW sex chromosomes, or if this species has a diverse array of sex determination mechanisms.
Mechanisms Responsible for the Differences in Karyotype between G. aculeatus and A. quadracus
Although G. aculeatus and A. quadracus diverged approximately 40 million years ago, our comparative FISH study has uncovered genomic rearrangements encompassing nearly half of the 21 G. aculeatus chromosome pairs. These rearrangements account for the 2 major karyotypic differences between these species: A. quadracus has a higher diploid number, coupled with a lower number of metacentric and submetacentric chromosomes, when compared to G. aculeatus (table 4).
The larger diploid number (46) in A. quadracus is explained by our finding that 2 G. aculeatus chromosome pairs (4 and 7) correspond to 4 A. quadracus chromosome pairs. Examination of other stickleback species does not reveal whether the G. aculeatus or A. quadracus diploid number is more similar to the ancestral state (fig. 1). The closest extant relative of G. aculeatus also has a diploid chromosome number of 42, as does a more distant relative, the ninespine stickleback, P. pungitius [Ocalewicz et al., 2008, 2011; Ross et al., 2009; Shapiro et al., 2009]. However, many differences in chromosome morphology exist between the Gasterosteus species and P. pungitius [Ocalewicz et al., 2008, 2011; Ross et al., 2009]. A recent report shows that P. pungitius has a higher NF:2n than G. aculeatus [Ocalewicz et al., 2011], indicating a higher number of metacentric and submetacentric chromosome pairs in P. pungitius. However, it is not known if the chromosome pairs that differ in morphology in G. aculeatus and P. pungitius are identical to the chromosome pairs that differ in G. aculeatus and A. quadracus in this study. The closest extant relative of P. pungitius, the brook stickleback (Culaea inconstans), has the same diploid number as A. quadracus, but these 2 species also differ in karyotype [Chen and Reisman, 1970; Ross et al., 2009]. No published reports document the diploid chromosome number and chromosome morphology of the fifteenspine stickleback (Spinachia spinachia), which is the closest extant relative of A. quadracus [Kawahara et al., 2009]. Thus, we lack comprehensive karyotype data from a sufficient number of stickleback species and populations to know whether the karyotype of G. aculeatus or A. quadracus is the more ancestral for this family.
Though the ancestral stickleback karyotype is unknown, our FISH screen has clarified some of the mechanisms that have shaped the A. quadracus and G. aculeatus karyotypes since they diverged. While metacentric and submetacentric chromosomes make up less than one quarter of the A. quadracus karyotype, they comprise more than one third of the G. aculeatus karyotype (table 4). One factor that contributes to this difference is the separation of G. aculeatus chromosomes 4 and 7 in the A. quadracus karyotype (fig. 2; table 3). The differential state of these chromosomes in G. aculeatus and A. quadracus, and the fact that they are both acrocentric in A. quadracus, suggest that Robertsonian fusions and/or centric fissions have played roles in the evolution of stickleback karyotypes. These fusions may leave signatures in the form of interstitial telomeric sequences (ITSs) in the species with the derived chromosome state. A recent study of a G. aculeatus population from Poland failed to detect any ITSs by FISH; however, it is possible that ITSs were eliminated or remain in a copy number that is below the detection threshold for FISH [Ocalewicz et al., 2011]. Thus, the ancestral state of chromosomes 4 and 7 in the stickleback family is still unknown, and proper determination of the ancestral state will clarify whether fusions or fissions are responsible for these differences in karyotype.
We also uncovered evidence for inversions involving at least 8 chromosomes, based on differences in relative centromere position and/or BAC probe hybridization positions between G. aculeatus and A. quadracus. Six of these inversion events encompass the centromere, and 5 of those inversions alter the gross chromosome morphology between G. aculeatus and A. quadracus. In particular, 3 of these 5 inversions are on chromosomes that are metacentric or submetacentric in G. aculeatus and acrocentric or telocentric in A. quadracus (table 3), which is one factor that accounts for the lower NF in this species compared to G. aculeatus (table 4). Future studies of chromosome morphology and relative probe hybridization positions in other stickleback species could further illuminate the role of pericentric inversions in the evolution of karyotypes across this family.
Conclusions
Our FISH survey has identified the major chromosome rearrangements that have shaped the G. aculeatus and A. quadracus karyotypes since they diverged approximately 35 million years ago. The A. quadracus karyotype contains a large number of acrocentric and telocentric chromosomes that can be explained by several pericentric inversions and Robertsonian fusion or centric fission events. Importantly, we discovered unexpected diversity among A. quadracus sex chromosomes. Male and female individuals from Connecticut do not harbor heteromorphic (distinguishable) sex chromosomes, in contrast to other A. quadracus populations in New England. Future studies are required to uncover the true diversity of sex chromosomes and sex determination mechanisms in this species and to learn how the diverse array of sex chromosome systems evolved in the stickleback family.
Acknowledgements
We thank Thomas Near for providing our wild-caught A. quadracus samples, the Broad Institute and Stanford Human Genome Center for allowing us to use BAC end sequences and the initial stickleback genome sequence assembly prior to publication, Joseph Ross for providing both protocols and training for stickleback cytogenetics and FISH, Kohta Yoshida for helpful discussions on chromosome evolution in fishes, and Anna Greenwood for her unique insights, discussions, and advice as this project progressed. We thank Barbara Trask, Joseph Ross, and Anna Greenwood for helpful comments and suggestions regarding this manuscript. Finally, we thank Stephen Uy, who translated key passages from Robert Matthey's Les Chromosomes des Vertébrés. Our work was funded by National Institutes of Health grants P50 HG002568 (C.L.P.), R01 GM089733 (C.L.P.), and T32 HG00035 (J.R.U.).
References
- Almeida-Toledo LF, Foresti F, Péquignot EV, Daniel-Silva MFZ. XX:XY sex chromosome system with X heterochromatinization: an early stage of sex chromosome differentiation in the Neotropic electric eel Eigenmannia virescens. Cytogenet Cell Genet. 2001;95:73–78. doi: 10.1159/000057020. [DOI] [PubMed] [Google Scholar]
- Almeida-Toledo LF, Daniel-Silva MFZ, Moysés CB, Fonteles SBA, Lopes CE, et al. Chromosome evolution in fish: sex chromosome variability in Eigenmannia virescens (Gymnotiformes: Sternopygidae) Cytogenet Genome Res. 2002;99:164–169. doi: 10.1159/000071589. [DOI] [PubMed] [Google Scholar]
- Andreata AA, Almeida-Toledo LF, Oliveira C, Toledo-Filho SA. Chromosome studies in Hypoptopomatinae (Pisces, Siluriformes, Loricariidae): I. XX/XY sex chromosome heteromorphism in Pseudotocinclus tietensis. Cytologia. 1992;57:369–372. [Google Scholar]
- Bertollo LAC, Born GG, Dergam JA, Fenocchio AS, Moreira-Filho O. A biodiversity approach in the Neotropical fish Hoplias malabaricus. Karyotype survey, geographic distribution of cytotypes and cytotaxonomic considerations. Chromosome Res. 2000;8:603–613. doi: 10.1023/a:1009233907558. [DOI] [PubMed] [Google Scholar]
- Bertollo LAC, Oliveira C, Molina WF, Margarido VP, Fontes MS, et al. Chromosome evolution in the Erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes) Heredity. 2004;93:228–233. doi: 10.1038/sj.hdy.6800511. [DOI] [PubMed] [Google Scholar]
- Chen TR, Reisman HM. A comparative chromosome study of the North American species of sticklebacks (Teleostei: Gasterosteidae) Cytogenetics. 1970;9:321–332. doi: 10.1159/000130102. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Bertollo LAC. Initial steps in XY chromosome differentiation in Hoplias malabaricus and the origin of an X1X2Y sex chromosome system in this group. Heredity. 2010;105:554–561. doi: 10.1038/hdy.2010.18. [DOI] [PubMed] [Google Scholar]
- Cioffi MB, Martins C, Bertollo LAC. Chromosome spreading of associated transposable elements and ribosomal DNA in the fish Erythrinus erythrinus. Implications for genome change and karyoevolution in fish. BMC Evol Biol. 2010;10:271–279. doi: 10.1186/1471-2148-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover DO, Heins SW. Adaptive variation in environmental and genetic sex determination in a fish. Nature. 1987;326:496–498. doi: 10.1038/326496a0. [DOI] [PubMed] [Google Scholar]
- Henning F, Moysés CB, Calcagnotto D, Meyer A, Almeida-Toledo LF. Independent fusions and recent origins of sex chromosomes in the evolution and diversification of glass knife fishes (Eigenmannia) Heredity. 2011;106:391–400. doi: 10.1038/hdy.2010.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornaday K, Alexander S, Breden F. Absence of repetitive DNA sequences associated with sex chromosomes in natural populations of the Trinidad guppy (Poecilia reticulata) J Mol Evol. 1994;39:431–433. [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 2009;50:401–404. doi: 10.1016/j.ympev.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Kingsley DM, Peichel CL. The molecular genetics of evolutionary change in sticklebacks. In: Östlund-Nilsson S, Mayer I, Huntingford FA, editors. Biology of the Three-Spined Stickleback. Boca Raton: CRC Press; 2007. pp. 41–82. [Google Scholar]
- Kingsley DM, Zhu B, Osoegawa K, De Jong PJ, Schein J, et al. New genomic tools for molecular studies of evolutionary change in threespine sticklebacks. Behaviour. 2004;141:1331–1344. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhardt M. Some aspects of karyoevolution in fishes. Anim Res Dev. 1998;47:7–36. [Google Scholar]
- Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Mattern MY. Molecular phylogeny of the Gasterosteidae: the importance of using multiple genes. Mol Phylogenet Evol. 2004;30:366–377. doi: 10.1016/s1055-7903(03)00190-8. [DOI] [PubMed] [Google Scholar]
- Matthey R: Les Chromosomes des Vertébrés (Librairie de L'université F. Rouge, Lausanne 1949).
- Moran P, Martinez JL, Garcia-Vazquez E, Pendas AM. Sex chromosome linkage of 5S rDNA in rainbow trout (Oncorhynchus mykiss) Cytogenet Cell Genet. 1996;75:145–150. doi: 10.1159/000134466. [DOI] [PubMed] [Google Scholar]
- Nanda I, Volff JN, Weis S, Korting C, Froschauer A, et al. Amplification of a long terminal repeat-like element on the Y chromosome of the platyfish, Xiphophorus maculatus. Chromosoma. 2000;109:173–180. doi: 10.1007/s004120050425. [DOI] [PubMed] [Google Scholar]
- Ocalewicz K, Fopp-Bayat D, Woznicki P, Jankun M. Heteromorphic sex chromosomes in the ninespine stickleback Pungitius pungitius. J Fish Biol. 2008;73:456–462. [Google Scholar]
- Ocalewicz K, Woznicki P, Furgala-Selezniow G, Jankun M. Chromosomal location of Ag/CMA3-NORs, 5S rDNA and telomeric repeats in two stickleback species. Ital J Zool. 2011;78:12–19. [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, et al. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader O, Ahne R, Fuchs J, Schubert I. Karyotype analysis of Helianthus annuus using Giemsa banding and fluorescence in situ hybridization. Chromosome Res. 1997;5:451–456. doi: 10.1023/a:1018412912790. [DOI] [PubMed] [Google Scholar]
- Schultheis C, Böhne A, Schartl M, Volff JN, Galiana-Arnoux D. Sex determination diversity and sex chromosome evolution in poeciliid fish. Sex Dev. 2009;3:68–77. doi: 10.1159/000223072. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Summers BR, Balabhadra S, Aldenhoven JT, Miller AL, et al. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr Biol. 2009;19:1140–1145. doi: 10.1016/j.cub.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN, Schartl M. Variability of genetic sex determination in poeciliid fishes. Genetica. 2001;111:101–110. doi: 10.1023/a:1013795415808. [DOI] [PubMed] [Google Scholar]
- White MJD. Modes of Speciation. San Francisco: W.H. Freeman and Company; 1978. [Google Scholar]
- Wootton RJ. The Biology of the Sticklebacks. London: Academic Press; 1976. [Google Scholar]