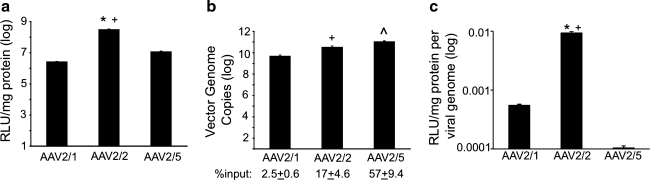
FIG. 1.
Recombinant adeno-associated virus (rAAV) transduction and entry profiles in HeLa cells. (a) Relative transduction efficiencies of the rAAV2/1, rAAV2/2, and rAAV2/5 vectors as assessed by luciferase expression. rAAV vectors pseudotyped with type 1, 2, or 5 capsids were used to transduce HeLa cells at a multiplicity of infection (MOI) of 1000 vector genome copies (VGC)/cell. The cells were infected for 24 hr, and luciferase assays were conducted on cell lysates. The relative light units (RLUs) in each sample were normalized to the total amount of protein in the lysate. Average values for each vector were calculated and plotted on a logarithmic scale (error bars=mean±SEM, n=4 independent experimental samples). (b) Relative entry efficiencies as assessed by virus genome content. rAAV-luciferase vectors were incubated with HeLa cells (MOI=1000 VGC/cell) at 4°C for 1 hour. Unbound virions were washed off with cold PBS, and the cells were further incubated with attached virus at 37°C for 1 hr to allow for virus entry. Cells were then collected by trypsinization, and post-nuclear supernatants (PNS) were isolated from cell homogenates. The total number of VGC in each sample was determined by TaqMan PCR and normalized to the value obtained for the corresponding vector in cells not shifted to 37°C. Average total VGC were calculated and plotted on a logarithmic scale (error bars=mean±SEM, n=3 independent experimental samples). The percent input was determined by calculating the ratio of viral genomes detected by TaqMan PCR to the total viral genomes applied to each well. (c) Efficiency of AAV transduction was calculated as the level of transduction (RLU/mg protein) divided by the VGC in cells at the time of harvest. The resulting average values were then plotted (error bars=mean±SEM, n=4 independent experimental samples). Statistical analysis was performed using one-way ANOVA with post-test (Tukey's multiple comparison): AAV2/1 vs. AAV2/2, *p<0.01; AAV2/2 vs. AAV2/5, +p<0.01; AAV2/1 vs. AAV2/5, ^p<0.01.