Figure 1.
Ribosome Profiling in Mouse Embryonic Stem Cells
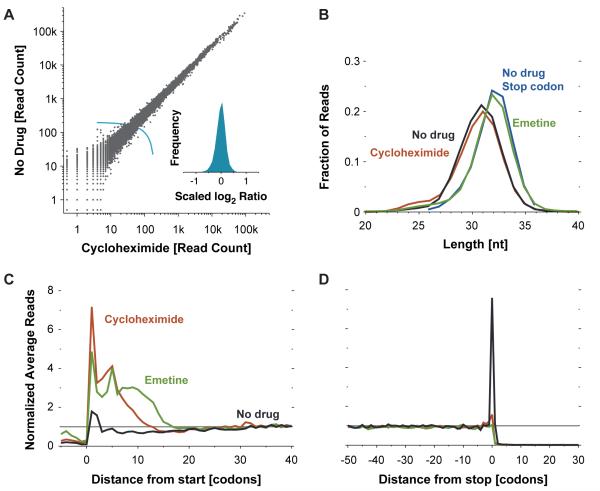
(A) Effect of elongation inhibitors on ribosome density. The number of ribosome footprint reads that align to the body of each coding sequence (Methods) is plotted for cells that were either untreated or pretreated with cycloheximide (Spearman r = 0.99). The inset shows a histogram of log2 ratios for genes with at least 200 total reads (the threshold shown by the light blue line) normalized by the median ratio (N = 10045, s.d. = 0.20, corresponding to 15 percent difference in measurements).
(B) Ribosome-protected fragment lengths. Plotted is the length distribution of ribosome footprints over the body of messages prepared from cells treated as indicated, as well as for footprints centered on the stop codon for the untreated cells.
(C) Metagene analysis of translation initiation. Average ribosome read density profiles over 4994 well-expressed genes (Table S1), aligned at their start codon, are shown for untreated and drug-treated samples.
(D) Metagene analysis of translation termination. As in (C) but alignment was from stop codons.