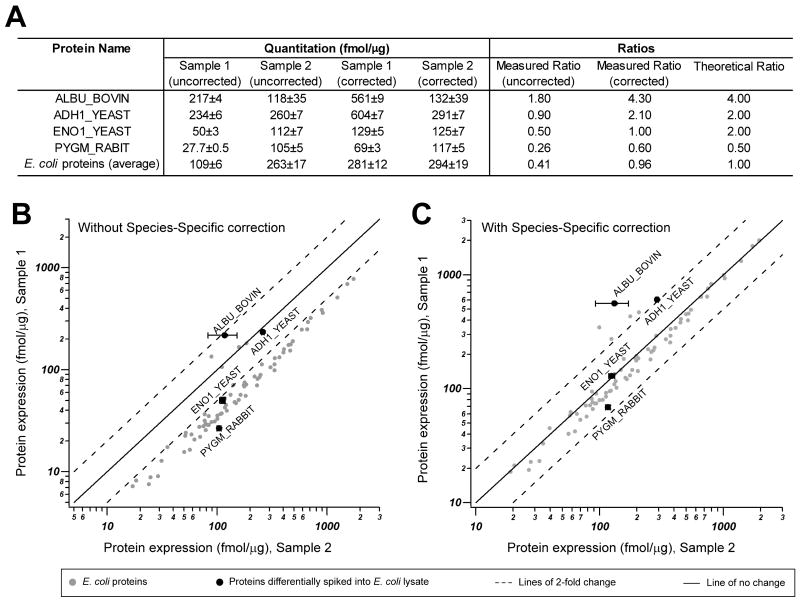
Figure 2. Validation of label-free protein quantification of a complex mixture of proteins from two different species (“mixed proteomes”).
A. We generated an artificial E. coli – mouse brain model system to assess the accuracy of MS-based label free quantification of proteins in a multi-species protein lysate. In addition, four exogenous proteins were spiked at pre-defined ratios as indicated (theoretical ratio) into 1:1 E. coli:mouse brain lysate (Sample 1, representing a “mixed proteome”) or E. coli lysates (Sample 2, representing a single proteome). After trypsin digestion and LC-MS/MS analysis, the quantitative measurements (fmol/μg) of proteins were assessed with and without species-specific correction. When species-specific correction was applied, the corrected measured ratio was almost identical to the theoretical ratio. B-C. Lower panels show a graphic representation of the direct quantitative comparison of all proteins from Sample 1 and Sample 2, without and with species-specific correction, as indicated. Note that when all proteins in the sample are considered and no correction is applied (B), an obvious quantitative bias towards lower quantities in Sample 1 by approximately 2× is observed, as expected. When using the total micrograms of only the identified E. coli proteins to normalize protein concentration as means of “species-specific” correction (C), the result is that the correct ratio of E. coli and spiked-in proteins are reproduced. ADH1_YEAST, yeast alcohol dehydrogenase; ENO1_YEAST, yeast enolas; ALBU_BOVIN, bovine albumin; PYGM_RABBIT, rabbit glycogen phosphorylase. A logarithmic scale was used for x and y axis.