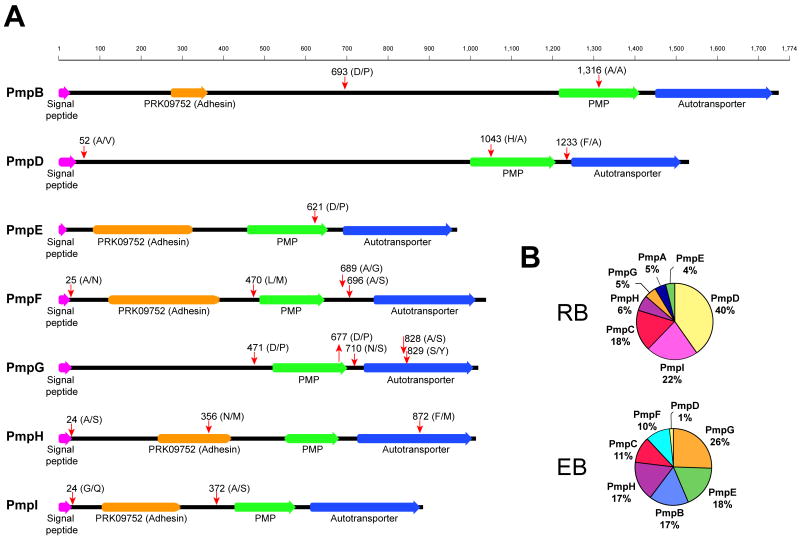
Figure 4. Semitryptic peptide analysis indicates that C. trachomatis polymorphic membrane proteins (PMPs) are extensively processed.
(A) Semitryptic peptides corresponding to PMPs were analyzed and mapped to the corresponding protein sequence. Potential cleavage sites identified are shown (red arrows). Cleavage motif, amino acid position, domains and signal peptides are shown. Signal peptides are indicated based on SignalP 3.0 prediction (http://www.cbs.dtu.dk/services/SignalP/) trained for Gram negative bacterial species. (B) Pie chart representing the relative abundance of different PMPs in the EB and RB forms, expressed as percentage of total mass corresponding to PMPs.