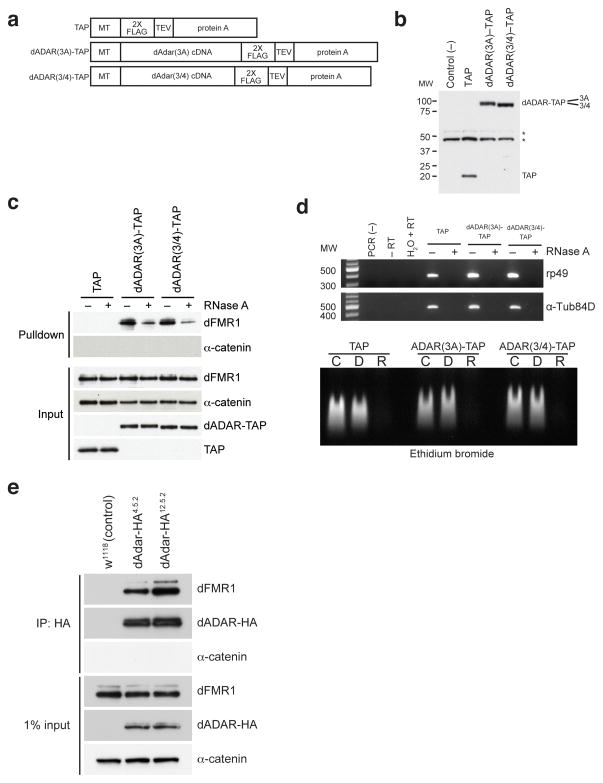
Figure 1. dFMR1 biochemically interacts with dADAR in Drosophila S2 cell culture and in vivo.
(a) Structure of TAP (consisting of 2X FLAG and protein A sequences separated by a TEV cleavage site), dADAR(3A)-TAP and dADAR(3/4)-TAP constructs used to generate stable S2 cell lines. Constructs are under control of an inducible metallothionein (MT) promoter. (b) Western analysis showing expression of constructs in transfected S2 cells. Untransfected S2 cells were used as a negative control for the FLAG antibody. Astericks denote non-specific bands present in all samples that were detected by the anti-FLAG antibody. Molecular weight (MW) on left is measured in kilodaltons (kDa). (c) Eluates from TAP pulldown followed by TEV cleavage show that dFMR1 associates with dADAR-TAP in the presence of RNase A. Samples treated or untreated with RNase A are designated as (+) or (−), respectively. α-catenin was used as a loading control and does not associate with dADAR-TAP. A FLAG antibody was used to detect TAP constructs in input lanes. (d) RT-PCR analysis (upper panel) and ethidium bromide staining of total RNA (lower panel) on RNase A-treated and control lysates, showing efficient RNA degradation in RNase-treated samples. For RT-PCR analysis (upper panel), samples treated or untreated with RNase A are designated as (+) or (−), respectively. Primers against ribosomal protein 49 (rp49) and α-Tubulin84D (α-Tub84D) were used for PCR amplification. Molecular weight marker (MW) denotes size migration in basepairs (bp). For ethidium bromide staining of total RNA (lower panel), total RNA from TAP, dADAR(3A)-TAP, and dADAR(3/4)-TAP lysates were treated with DNase I (D), RNase A (R) or were untreated (C). (e) Co-IP experiments performed on head lysates prepared from w1118 (control) and two independent endogenously tagged dADAR-HA fly lines, dAdar-HA4.5.2 and dAdar-HA12.5.2. An HA antibody was used to detect dADAR-HA and α-catenin was used as a loading control and negative control for the co-IP.