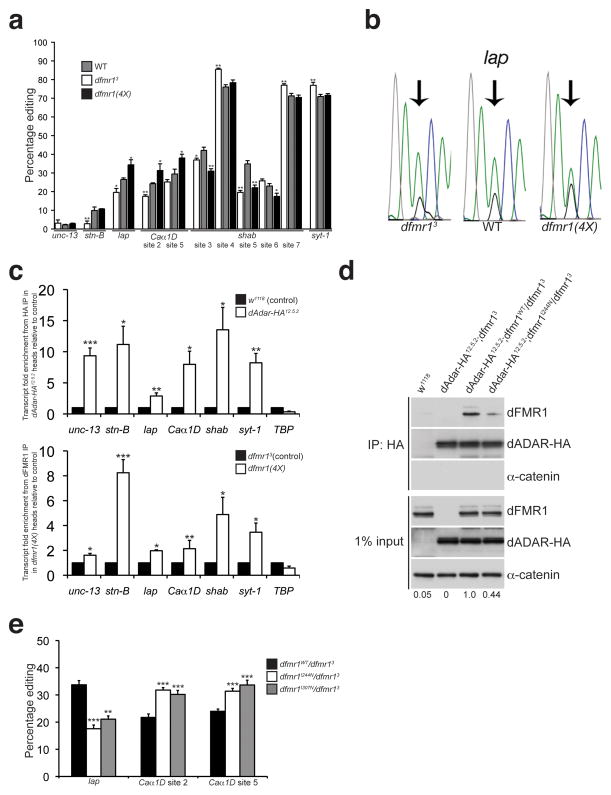
Figure 7. dFMR1 modifies dADAR function and affects A-to-I editing efficiency.
(a) Percentage of editing observed in samples from WT (gray bars), dfmr13 (white bars), and dfmr1(4X) (black bars) larvae was quantified and graphed for the following transcripts: unc-13, stoned-B (stn-B), lap, Caα1D, shab, and synaptotagmin-1 (syt-1). n = 3–7 individual RT-PCR reactions for each site. *p<0.05, **p<0.01, WT vs. dfmr13 and WT vs. dfmr1(4X) were analyzed with Mann Whitney-U test. Error bars denote s.e.m. (b) Representative electropherograms for the lap edited adenosine sequenced from WT, dfmr13, and dfmr1(4X) whole larval cDNA. Arrows point to edited sites analyzed. Green peak represents unedited (A) site, and black peak represents edited (G) site. (c) dADAR and dFMR1 associate with edited transcripts in vivo. RNA immunoprecipitation of transcripts associating with dADAR-HA or dFMR1 in adult head lysates. Fold enrichment of transcripts in dAdar-HA12.5.2 samples relative to w1118 was performed for the dADAR-HA RNA immunoprecipitation in the upper graph, and fold enrichment of transcripts in dfmr1(4X) overexpressing flies relative to dfmr13 null flies is shown for the dFMR1 RNA immunoprecipitation (lower graph). TBP was used as an unedited, non-specific transcript. Quantification of transcript enrichment was performed by using quantitative RT-PCR and fold enrichment was normalized to fold change of actin mRNA. Results represent four independent immunoprecipitation experiments for each HA and dFMR1 RNA IP and quantitative RT-PCR experiments was performed using three technical replicates. *p<0.05, **p<0.01, ***p<0.001, analyzed with Student’s t-test. Error bars denote s.e.m. (d) A mutation in the KH1 RNA binding domain of dFMR1 reduces the robustness of the dFMR1:dADAR biochemical interaction. co-IP experiments were performed with flies containing a wild-type dFMR1 construct (dfmr1WT) or a construct containing a point mutation in the dFMR1 KH1 domain (dfmr1I244N) that were crossed to the dAdar-HA12.5.2;dfmr13 fly line. w1118 flies served as a negative control for the HA antibody. An HA antibody was used to detect dADAR-HA and α-catenin was used as a loading control and negative control for the co-IP. Expression levels of dFMR1 in the IP samples were normalized to dFMR1 input levels and average fold change relative to dAdar12.5.2;dfmr1WT/dfmr13 is denoted below each lane. Experiment was performed three times. (e) Mutations in the dFMR1 KH domains affect editing of lap and Caα1D. Percentage of editing observed in dfmr1WT/dfmr13 (black bars), dfmr1I244N/dfmr13 (white bars), and dfmr1I307N/dfmr13 (gray bars) whole larvae. n = 3 8 individual RT-PCR reactions for each site. **p<0.01, ***p<0.001, dfmr1WT/dfmr13 vs. dfmr1I244N/dfmr13 and dfmr1WT/dfmr13 vs. dfmr11307N/dfmr13 were analyzed with Mann Whitney-U test. Error bars denote s.e.m.