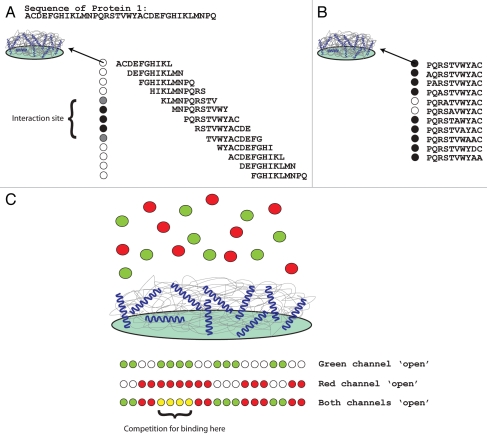
Figure 1.
Using peptide arrays to study protein-protein interactions. (A) When 2 interacting proteins have been identified, the interaction site of one of the proteins on the other can be identified if one of the proteins is available in a purified recombinant form. To do this, protein 1 is synthesized on a cellulose support using SPOT synthesis. The protein is made in short overlapping peptides (in this figure, a simulated protein sequence is shown which is then synthesized as a set of overlapping 10-mer peptides which are shifted to the right by 2 amino acids). When overlayed with recombinant protein 2, the binding site of the overlayed protein on the immobilized protein (peptides) will be revealed. (B) To refine the binding site and identify the specific amino acids in Protein 1 required for the interaction of protein 2, an alanine substitution peptide array is synthesized. To do this, a parent peptide (or several) is chosen from the first screen (A), in this case; PQRSTVWYAC from protein 1. New immobilized peptides are synthesized but in this case, each peptide differs from the parent peptide by 1 amino acid (each amino acid is subsequently changed to an alanine, alanines are changed to aspartic acid). The array is overlayed with protein 2 as described above, and the critical amino acids required for the interaction are revealed (ST from the example shown). (C) When 2 proteins are suspected to compete for binding to another protein, both competing proteins can be overlayed in equal concentration on an immobilized protein (peptide). One protein can be conjugated to a green dye, one protein can be conjugated to a red dye. In combination with an Odyssey® infrared imager (LI-COR Biosciences) competition between two proteins for binding to the same immobilized protein (peptide) can be investigated. ‘Green spots’ indicate a binding preference for protein 1, ‘red spots’ indicate a binding preference for protein 2 and ‘yellow spots’ indicate that the two proteins compete for binding to the immobilized peptide sequence.