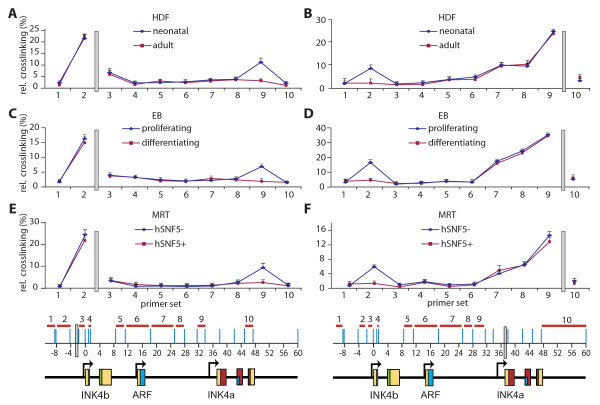
Figure 4.
Regulated chromatin looping between INK4a and INK4b. (A-F) Chromatin conformation capture - quantitative polymerase chain reaction (qPCR) analyses of long-range interactions at the INK4b-ARF-INK4a locus. Locus-wide relative cross-linking frequencies of EcoRI fragments 1 to 10 are plotted. The constant primer and the TaqMan probe (grey bar) were either in the EcoRI fragment -4 to -2 kb upstream of INK4b (A, C and E) or overlapping the INK4a promoter (B, D and F). The locations within the INK4b-ARF-INK4a genomic region of the EcoRI sites (blue lines), bait fragment (harbouring the grey bar, indicating the TaqMan probe) and suitable candidate EcoRI fragments (numbered red segments) are shown at the bottom. The relative cross-linking frequency of each fragment is plotted, with the fragment numbers indicated on the X-axis. (A, B) Neonatal (blue) and adult (red) human diploid fibroblasts. (C, D) Proliferating- (blue) and differentiating (red) eryhtroblasts. (E, F) Proliferating MRT cells lacking hSNF5 (blue) and senescing MON cells expressing hSNF5 (red). Each point represents the mean of three independent biological replicates, each analysed in triplicate by qPCR. Standard deviations are depicted by error bars.