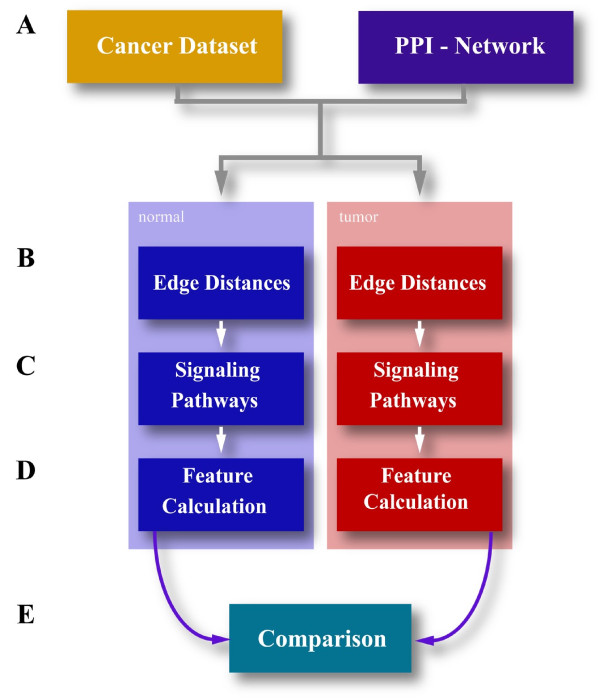
Figure 5.
Workflow of the method. (A) Gene expression data from normal and tumor samples was mapped onto the respective nodes of the protein-protein-interaction network. (B) Node distances dxy were calculated from correlation coefficients of neighboring genes in the network for normal and tumor samples resulting in one normal and one tumor network with weighted links. Transcription factors and receptors were selected from public data repositories (Gene Ontology and TRANSFAC). (C) Shortest paths were calculated for all pair-wise combinations of receptors and transcription factors. Links and nodes that did not appear in any shortest path were removed and the largest connected component of the remaining network was used as the representative signaling network. (D) Network features were calculated for each signaling network and (E) the results for the networks of tumor and normal samples compared.