Figure 1.
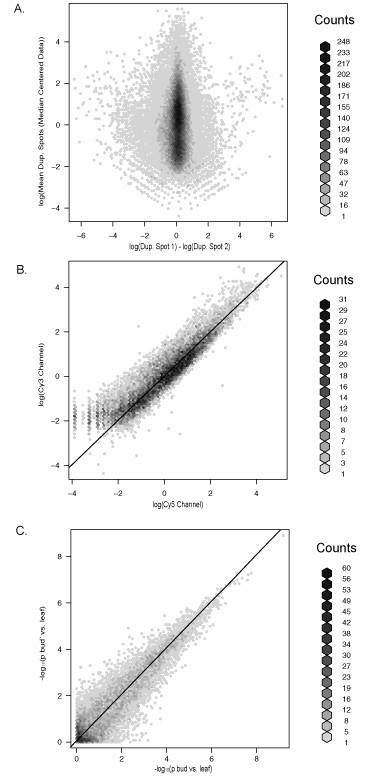
A reproducible platform for transcription profiling using cotton microarrays. A) Differences detected between duplicated spots plotted by their mean expression value from replicate 1 of the experiment. Each plot is hexagonally-binned to uncover the density component that is otherwise saturated by a cloud of data points. The difference of the log-adjusted, median centered duplicate spots is measured on the horizontal axis and the mean value of the same duplicated spots is measured on the vertical (values in grey, scale bars on left vertical axis). Most duplicated spots deviated very little, though genes with lower expression values tended to deviate more. Replication of the three treatment loop design indicated only minor detectable differences between duplicated spots. Nearly identical results were found for the other two replicates. B) Correlation of normalized, log-adjusted fluoresce intensity values for bud (Cy3) × bud' (Cy5) for the first microarray of the first replication. A 45° angle line has been overlaid to illustrate the expected 1:1 ratio of spot intensity. In this case, Cy3 labeled aRNA had higher intensity values on average; however, the effect of dye was removed from the contrast of differential expression in our analysis by including a dye component into our analytical model through dye swaps. C) Correlation of t-test p-values. Gene-specific tests for differential expression between bud and leaf and between bud' and leaf were conducted as described in Materials and Methods. The p-values from the bud' vs. leaf tests are plotted against the p-values from the bud vs. leaf tests on a negative natural log scale. The points in the upper right quadrant of the picture correspond to the genes with the smallest p-values. The points are scattered tightly around the 45 degree line, indicating that that p-values for the most significant genes were very similar according to both comparisons.
