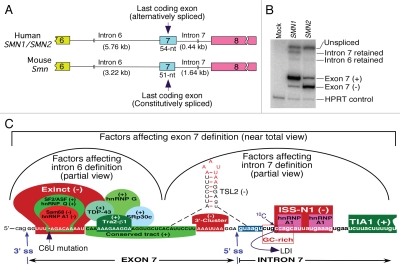
Figure 1.
Organization of human (SMN) and mouse (Smn) genes covering the last 3 exons. (A) Comparison of exon 7 and introns 6 and 7 of SMN and Smn genes. Exons are shown as colored boxes, whereas introns are shown as broken lines. Sizes of introns and exon 7 are given. (B) Results of RT-PCR for transcripts derived from SMN1 and SMN2 minigenes.26 Note the presence of intron 7 retained intermediate due to a weak 5′ ss in SMN1. As per intron definition model, splicing of SMN1 exon 7 was mainly driven by intron 6 definition due to a strong 3′ ss. C6U mutation causes a loss on intron 6 definition in SMN2 leading to skipping of exon 7. (C) Partial view of factors responsible for intron definition. In vivo selection of entire SMN exon 7 revealed three major regulatory regions: Exinct, conserved tract and 3′-cluster.26 SF2/ASF, hnRNP A1, Sam68 and hnRNP Q interact within the region corresponding to Exinct.40,41,45,47 Binding site of tra2-β1 and associated factors hnRNP G, SRp30c and TDP-43 fall within conserved tract.49–52 The 3′-cluster falls within the terminal stem loop 2 (TSL2) structure that also sequesters the 5′ ss of exon 7.54 ISS-N1 is a major intronic inhibitory elements that harbors 2 hnRNP A1 motifs.27,56 ISS-N1 also overlaps with a GC-rich sequences that provides the shortest antisense target for correction of SMN2 exon 7 splicing in SMA.55 The cytosine residue at the 10th intronic position (10C) has been implicated in a long-distance interaction (LDI) with deep intronic sequences.57 TIA1 binds to an intronic region downstream of ISS-N1.25 Cis-elements and factors that are likely to define either intron 6 or intron 7 have been grouped. (+) and (−) indicates that a given cis-element or splicing factor promotes and suppresses exon 7 inclusion, respectively. Exon 7 sequences are shown in capital letters, whereas intronic sequences are shown in small case letters.