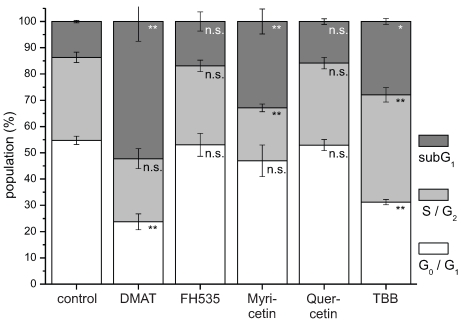
Figure 5.
Cell cycle analysis. CCLP-1 cells were incubated with either 10 µM DMAT, 20 µM FH535, 50 µM myricetin, 50 µM quercetin or 10 µM TBB in serum-free DMEM and incubated for 48 hrs. Analysis of cell cycle distribution was performed using ethanol-fixed, RNase-treated and PI-stained cells by flow cytometry. The percentage of cells in G0G1, S/G2, or below G0G1 (subG1 fraction) cell cycle phases was analysed with the FlowMax software (Partec, Görlitz, Germany). The subG1 fraction is a measure of apoptotic cell death as these cells have undergone apoptotic DNA fragmentation. Significant (p<0.05) and highly significant (p<0.01) differences between population sizes of treated cells and the untreated controls for each cell cycle phase are marked with * and **, respectively (univariate ANOVA, LSD post-hoc test).