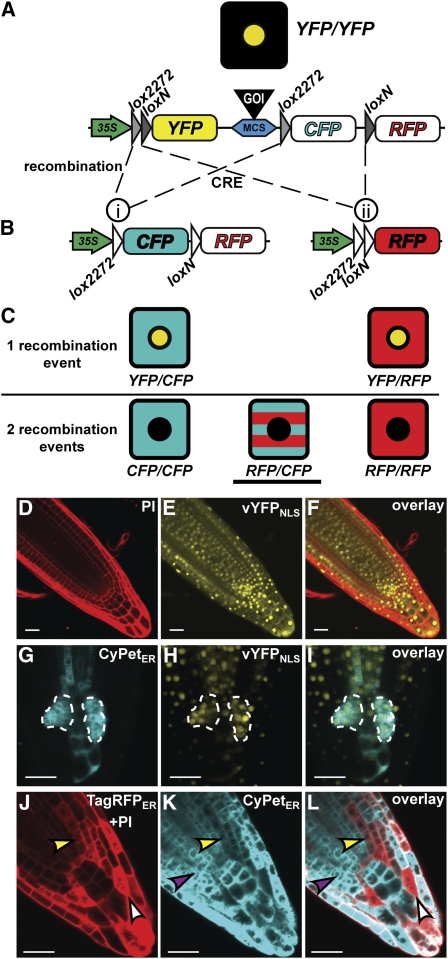
Figure 1.
Induction and Analysis of Clones Using the BOB System.
(A) and (B) The BOB construct before (A) and after (B) CRE-mediated recombination as indicated by dashed lines. Recombination between lox2272 sites (i) fuses the 35S promoter with CypetER. A transcriptional terminator (data not shown) at the 5′ of CyPetER prevents TagRFPER expression. Recombination between loxN sites fuses the 35S promoter to TagRFPER (ii). Colored, filled objects represent active 35S promoter (green), transcribed fluorescent proteins (yellow, cyan, and red), lox sites (gray triangles), and a multicloning site (blue) for cloning a GOI (black). White filled objects represent nonactive modules (i.e., CyPetER and TagRFPER before recombination and lox sites after recombination).
(C) Illustration of possible fluorescently marked cells resulting from one recombination (top) or two recombination events (bottom). Underlined cell depicts the type of clones that were used to identify NHCs in all analyses.
(D) to (F) Confocal images of BOB-RBR roots prior to CRE induction. Visualization of PI-marked cell walls in the red channel (D), vYFPNLS expression prior to CRE induction in the yellow channel (E), and their overlay (F).
(G) to (I) Confocal images of a root tip with two highlighted single recombination clones (dashed enclosure) visualized 5 d postinduction. Highlighted clones analyzed in the cyan channel (CyPetER; [G]), the yellow channel (vYFPNLS; [H]), and in the overlay (I).
(J) to (L) Efficient clone formation using the BOB system upon 1-h HS induction. Clones in a HS:CRE;BOB root tip 3 d after HS induction are shown in the red channel (PI + TagRFPER; [J]), the cyan channel (CyPetER; [K]), and overlay (double fluorescence; [L]). Representative CyPetER, TagRFPER, and double fluorescence clones are denoted by purple, white, and yellow arrowheads, respectively. Bars = 20 μm.