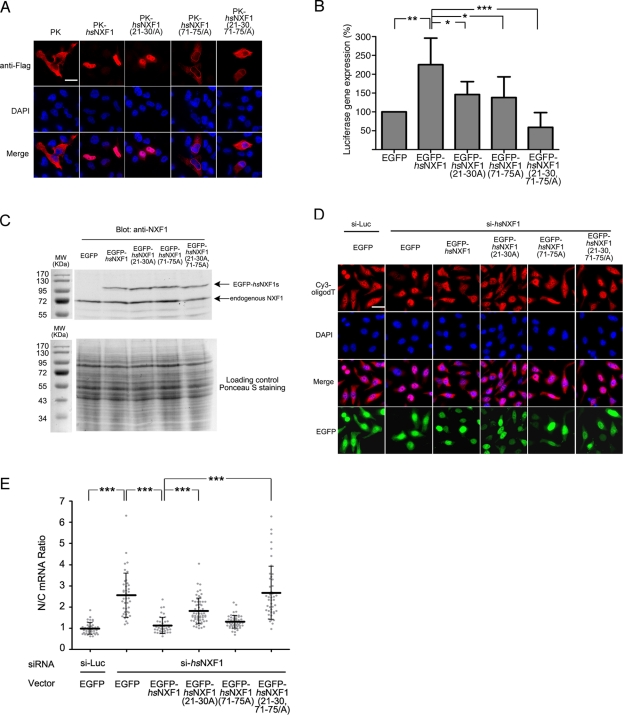
FIGURE 3:
NLS mutations impair nuclear localization of hsNXF1 and its ability to activate luciferase gene expression and export mRNA. (A) FLAG-tagged pyruvate kinase (PK)–hsNXF1 and its NLS mutants were transfected into HeLa cells. Localization of PK fusion proteins were detected by deconvolution microscope. Scale bars, 10 μm. (B) Luciferase reporter gene expression assays of hsNXF1 and its NLS mutants. EGFP-hsNXF1 proteins were cotransfected with pCMV-Luc vector, and the expression levels of the luciferase gene were calculated by normalizing the luciferase signals that were detected by Luciferase Assay System to CellTiter-Glo signals. The results are averages of six independent experiments ± SD. *p < 0.05; **p < 0.01; ***p < 0.001. (C) Expression levels of EGFP-hsNXF1 proteins and endogenous hsNXF1. (D) Wild type and mutant of pEGFP-C1-hsNXF1 vectors were cotransfected with siRNA targeting hsNXF1 for 48 h, and the localization of mRNA was detected by Cy3-oligodT. Scale bar, 10 μm. (E) Quantitation of C; the average nucleus/cytoplasm (N/C) ratios of Cy3 signals in ∼50 transfected cells from each group are shown as middle horizontal bars ± SD. Gray dots, data points. ***p < 0.001.