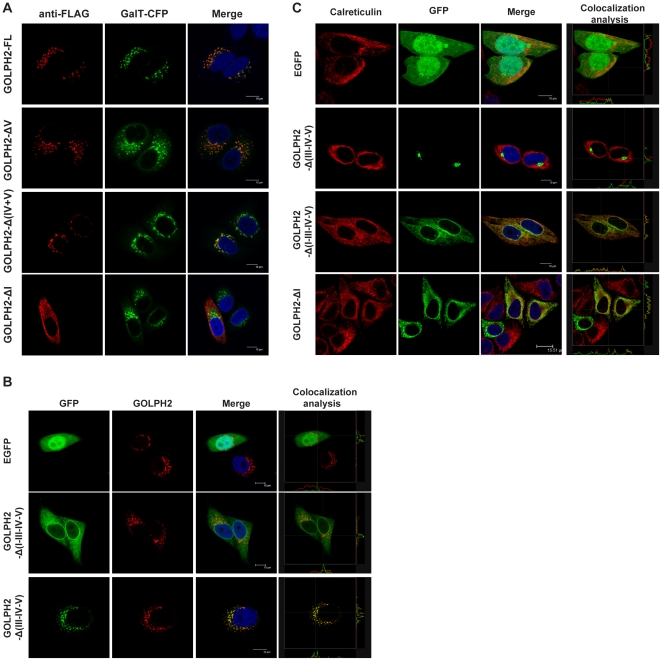
Figure 3. The TMD and cytoplasmic tail of GOLPH2 determine its Golgi localization.
A. Subcellular localization of GOLPH2 truncation proteins in HeLa cells. Recombinant proteins transiently expressed in HeLa cells were stained with anti-FLAG mAb, which emits red fluorescence. Subcellular localization was imaged using a confocal microscope. β-1,4-galactosyltransferase (GalT) fused with cyan fluorescent protein was cotransfected as a Golgi localization marker (shown as a green pseudocolor). Merged images show the colocalization of GOLPH2 truncation proteins and GalT. Bars, 10 µm. B. The TMD and cytoplasmic tail of GOLPH2 were sufficient for Golgi localization. HeLa cells were transfected with GOLPH2-Δ(III–IV–V), GOLPH2-Δ(I–III–IV–V), and pEGFP. About 24 h after transfection, cells were treated for 4 h with cycloheximide (100 µg/mL) to inhibit protein synthesis prior to fixation and permeabilization. Subcellular localization was imaged using a confocal microscope. Endogenous GOLPH2 was probed using anti-GOLPH2 mAb as a Golgi localization marker (red fluorescence). Merged images were analyzed using the Leica confocal software. Bars, 10 µm. C. Deletion of region I resulted in ER localization. GOLPH2-Δ(III–IV–V), GOLPH2-Δ(I–III–IV–V), GOLPH2-ΔI, and pEGFP were transfected into HeLa cells. Subcellular localization was imaged using a confocal microscope. Calreticulin was probed as an ER marker protein (red fluorescence).