SUMMARY
Autism spectrum disorders such as Rett syndrome (RTT) have been hypothesized to arise from defects in experience-dependent synapse maturation. RTT is caused by mutations in MECP2, a nuclear protein that becomes phosphorylated at S421 in response to neuronal activation. We show here that disruption of MeCP2 S421 phosphorylation in vivo results in defects in synapse development and behavior, implicating activity-dependent regulation of MeCP2 in brain development and RTT. We investigated the mechanism by which S421 phosphorylation regulates MeCP2 function and show by chromatin immunoprecipitation-sequencing that this modification occurs on MeCP2 bound across the genome. The phosphorylation of MeCP2 S421 appears not to regulate the expression of specific genes; rather, MeCP2 functions as a histone-like factor whose phosphorylation may facilitate a genome-wide response of chromatin to neuronal activity during nervous system development. We propose that RTT results in part from a loss of this experience-dependent chromatin remodeling.
INTRODUCTION
Mutations in MECP2 cause Rett syndrome (RTT), a human neurodevelopmental disorder that can lead to cognitive impairment, autistic features, motor disabilities, seizures, and anxiety (Chahrour and Zoghbi, 2007). Experiments that disrupt MeCP2 expression in specific populations of cells in mice indicate that dysfunction of neurons throughout the central nervous system contributes to the symptoms associated with RTT (Guy et al., 2010). However, the molecular mechanism by which disruptions in MeCP2 function give rise to neuronal dysfunction remains poorly understood.
MeCP2 was identified based on its affinity for methylated cytosines within DNA (Lewis et al., 1992). Because DNA methylation has been associated with transcriptional inhibition and because MeCP2 contains a domain that can mediate transcriptional repression in vitro (Nan et al., 1997), it was suggested that MeCP2 functions as a repressor of gene expression. Disruption of MECP2 in RTT was predicted to lead to the up-regulation of target genes, the identification of which might then be expected to provide insight into the etiology of this disorder. While both genetic and molecular biological approaches have been used to identify genes whose expression is regulated by MeCP2, the changes in gene expression detected when MeCP2 function is disrupted are small in magnitude, and are not entirely consistent with the idea that MeCP2 is a transcriptional repressor of specific genes (Chahrour et al., 2008; Tudor et al., 2002; Yasui et al., 2007). In conjunction with recent evidence that MeCP2 binds broadly throughout the genome and affects nucleosome structure (Ghosh et al., 2010; Skene et al., 2010), these findings suggest that, rather than acting as a sequence-specific transcription factor, MeCP2 functions as a global regulator of chromatin structure. However, it remains to be determined how the loss of MeCP2 function in the nucleus of neurons leads to a disruption of chromatin architecture, and why this gives rise to the defects associated with RTT.
The clinical course of RTT has provided clues as to the role of MeCP2 in the maturation of the nervous system. RTT-associated neuronal dysfunction first manifests itself in early postnatal life, when sensory experience is required for the refinement of developing neuronal circuits. This observation suggests that MeCP2 might mediate some of the effects of experience on synapse and neural circuit development, and that the absence of activity-dependent regulation of MeCP2 in RTT contributes to the etiology of this disorder.
In support of this idea, MeCP2 becomes newly phosphorylated at a specific amino acid residue, serine 421 in the brain in response to sensory stimuli (S421 refers to mouse MeCP2 isoform 2 and corresponds to S438 in mouse MeCP2 isoform 1 and S423 in human MeCP2) (Deng et al., 2010; Murgatroyd et al., 2009; Zhou et al., 2006). In cultured neurons, MeCP2 S421 phosphorylation is triggered by the release of glutamate at excitatory synapses, suggesting that synaptic activation may regulate MeCP2 function as part of an adaptive response to neuronal stimulation (Chen et al., 2003; Zhou et al., 2006). The phosphorylation of MeCP2 at S421 has been suggested to play a role in the neuronal activity-dependent induction of brain derived neurotrophic factor (BDNF), a secreted protein that promotes many aspects of experience-dependent synaptic development. Consistent with this possibility, when MeCP2 is over-expressed in neurons the phosphorylation of S421 affects the development of neuronal dendrites and spines (Zhou et al., 2006). The phosphorylation of S421 in response to neuronal activity has also been suggested to reduce the binding of MeCP2 to methylated DNA (Chen et al., 2003; Martinowich, 2003), leading to the hypothesis that synaptic activity regulates neuronal development by decreasing the affinity of MeCP2 for methylated cytosines within the regulatory regions of genes such as Bdnf, resulting in a change in chromatin structure that is required for gene activation.
The hypothesis that a disruption of activity-dependent gene regulation gives rise to the synaptic defects associated with RTT provides an attractive means to explain why identifying MeCP2 target genes under basal conditions has proven so challenging. However, all of the data that implicate MeCP2 phosphorylation in experience-dependent transcription and neuronal development stem from experiments in which MeCP2 was studied in vitro, using over-expression assays. It remains to be determined whether activity-dependent regulation of MeCP2 is required for brain development and if deficits in this process are sufficient to explain the phenotypes observed in mouse models of RTT.
To investigate the importance of MeCP2 S421 phosphorylation for the development of the nervous system and the pathogenesis of RTT, we generated a mouse in which MeCP2 S421 is converted to an alanine to prevent phosphorylation of this residue. We find that loss of MeCP2 S421 phosphorylation in vivo results in defects in dendritic and synaptic development and in abnormal behavioral responses to novel experience, suggesting that RTT is at least in part a disorder of experience-dependent brain development. We investigated the mechanism by which S421 phosphorylation regulates MeCP2 function and show by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-Seq) that this modification occurs on MeCP2 bound across the genome. The phosphorylation of MeCP2 S421 appears not to control the expression of specific genes; rather, MeCP2 functions as a histone-like factor whose phosphorylation regulates a genome-wide response of chromatin to neuronal activity during nervous system development. These findings suggest that aspects of RTT result from a loss of this experience-dependent chromatin remodeling.
RESULTS
Mutation of MeCP2 S421 Preserves Key Features of MeCP2 Protein Function
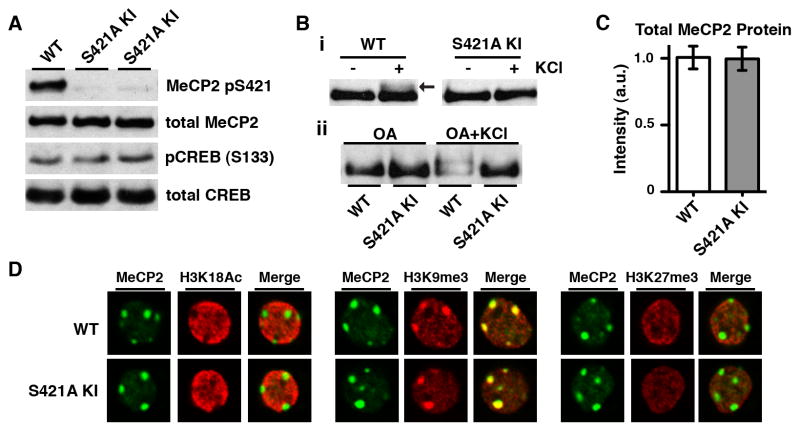
To examine how neuronal activity-dependent MeCP2 phosphorylation regulates brain development, we generated a knock-in mouse in which S421 of MeCP2 is mutated to an alanine (S421A) (Figure S1 available online). We reasoned that disruption of MeCP2 S421 phosphorylation in vivo might provide insight into how loss of activity-dependent MeCP2 regulation contributes to RTT. Western blotting and immunohistochemistry using an antibody that specifically recognizes MeCP2 when phosphorylated at S421 confirmed that MeCP2 S421 phosphorylation is eliminated in MeCP2 S421A mice (Figures 1A and S1F). Membrane depolarization of cultured cortical neurons by treatment with high extracellular potassium chloride (KCl) to induce calcium influx into neurons induces a slowly-migrating form of MeCP2 on an SDS-polyacrylamide gel that we have previously shown is due to phosphorylation at S421 (Chen et al., 2003; Zhou et al., 2006). This slowly-migrating form of MeCP2 was not induced by membrane depolarization of cultured neurons derived from the MeCP2 S421A knock-in mice, confirming that S421 has been converted to an amino acid that cannot be phosphorylated (Figure 1B).
Figure 1. Selective Loss of the Activity-Dependent Phosphorylation of MeCP2 S421 in vivo Preserves the Expression Level and Localization of MeCP2.
(A) Western blot analysis of nuclear extracts prepared from the forebrains of two 4-week-old male MeCP2 S421A knock-in mice (S421A KI) and their wild-type (WT) littermate using antibodies specific to MeCP2 pS421, and total MeCP2. Parrallel blots for pS133 CREB and total CREB show that there is no gross disruption of activity-dependent signaling in the brain.
(B) Anti-total MeCP2 Western blots of lysates from E16 + 5 DIV dissociated hippocampal neurons derived from MeCP2 S421A knock-in mice or wild-type littermates. (i) Arrow indicates the S421 phosphorylation-dependent slowly-migrating form of MeCP2 induced by 60 minute 55 mM KCl stimulation in wild-type cells (ii) 30 minute pre-treatment with okadaic acid (0.25 μM) to inhibit protein phosphatase activity increases the proportion of slowly-migrating MeCP2 induced by the 60 minute membrane depolarization. MeCP2 S421A neurons do not display this slowly-migrating form of MeCP2 in response to membrane depolarization.
(C) Quantification of Western blot analysis for the forebrains of 4-week-old MeCP2 S421A mice (n=3) and their wild-type littermates (n=3) using anti-total MeCP2 antiserum. Data are mean ± SEM. See also Figure S1.
(D) Immunocytochemistry of E16 + 22 DIV cortical neurons derived from MeCP2 S421A mice and wild-type littermates using an antibody against total MeCP2 and costaining with anti-histone H3 lysine 18 acetylation (H3K18Ac, active chromatin), anti-histone H3 lysine 9 trimethylation (H3K9me3, heterochromatin) or anti-histone H3 lysine 27 trimethylation (H3K27me3, repressed chromatin). Single 63X confocal planes of representative neuronal nuclei are shown.
Precise regulation of MeCP2 protein levels is required for proper nervous system function in humans and mice: both MECP2 gene duplications in humans and two-fold over-expression of MeCP2 in mice result in a spectrum of symptoms that overlap with those seen upon MeCP2 loss-of-function (Guy et al., 2010). Thus our ability to draw conclusions as to the importance of MeCP2 S421 phosphorylation in vivo depends critically on the maintenance of normal MeCP2 protein levels in MeCP2 S421A mice. Quantitative Western blotting revealed that total MeCP2 levels in the brains of MeCP2 S421A knock-in mice and their wild-type littermates are indistinguishable across a number of developmental time points (Figures 1C and S1G). Recent evidence indicates that MeCP2 is expressed in both neuronal and glial cells in the mouse brain (Ballas et al., 2009; Maezawa et al., 2009). We used immunocytochemistry in cultured cortical neurons to specifically determine if MeCP2 levels in neurons are perturbed by the S421A mutation. The expression of MeCP2 protein, as indicated by the intensity of anti-MeCP2 immunoreactivity, is similar in wild-type and MeCP2 S421A cortical neurons (Figure 1D). These findings indicate that the MeCP2 S421A mutation does not change the level of MeCP2 protein expression in the brain.
MeCP2 associates predominantly with heterochromatic foci in mouse cell nuclei in a DNA methylation-dependent manner, resulting in a characteristic pattern of diffuse MeCP2 staining throughout the nucleus, with prominent foci co-localizing with pericentromeric heterochromatic regions (Nan et al., 1996). We observed this pattern of staining with anti-MeCP2 antibodies in neurons derived from both wild-type and MeCP2 S421A mice (Figure 1D), indicating that mutation of MeCP2 S421 does not disrupt the subnuclear distribution of MeCP2 in the nucleus and suggesting that activity-dependent MeCP2 S421 phosphorylation may not be required for targeting of MeCP2 to chromatin. While it remains possible that the conservative mutation of S421 to alanine affects an undetected function of MeCP2 in addition to eliminating activity-dependent phosphorylation of this residue, we conclude that this mutation does not significantly alter levels of MeCP2 expression or nuclear localization of MeCP2. Given that the S421A mutation does not affect these fundamental properties of the MeCP2 protein, we reasoned that it should be possible to employ MeCP2 S421A knock-in mice to investigate the specific role of MeCP2 S421 phosphorylation during brain development.
MeCP2 S421 Phosphorylation Regulates Dendritic Development in vitro and in vivo
A major determinant of synapse formation and the integration of neurons into a circuit is the pattern of dendritic arborization receiving afferent input. Sensory-evoked neuronal activity has been shown to stabilize connections between neurons through the modulation of dendritic growth and patterning. While previous studies have implicated MeCP2 in dendritic growth both in vitro and in vivo, it has not been possible to determine if the absence of MeCP2 phosphorylation contributes to the defects in dendritic growth that occur in RTT.
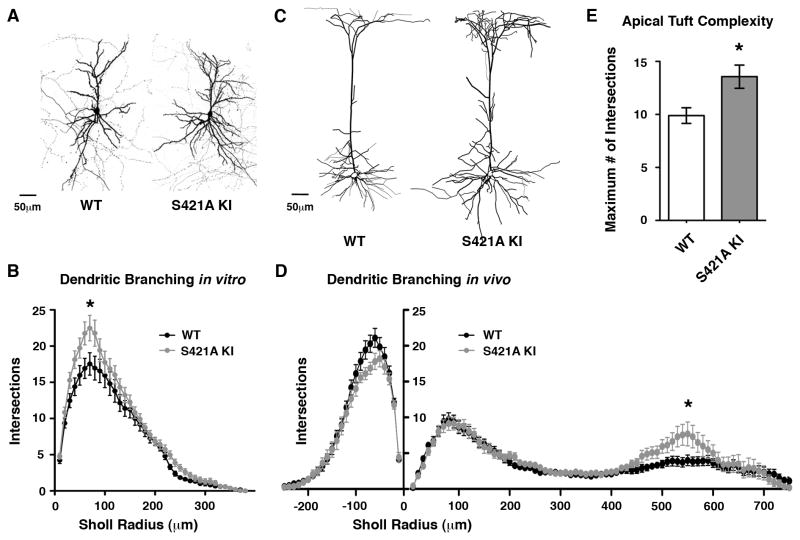
To investigate this possibility, we cultured cortical neurons from the brains of wild-type and MeCP2 S421A mice and monitored dendritic growth in these cultures. Sparse transfection of GFP allowed visualization of the dendritic arbors of individual cells, and cells with pyramidal morphology were imaged for analysis of dendritic patterning at DIV 21–22. The Sholl method of measuring dendritic complexity at a series of radii of increasing distances from the cell soma revealed a significant increase in the average number of dendritic branches in the MeCP2 S421A mutant (Figure 2A and 2B). We conclude that in cultured cortical neurons, MeCP2 S421 phosphorylation is required for proper dendritic development. Calcium signaling pathways initiated by neuronal activity influence both the net growth of dendritic arbors as well as the refinement of dendritic branching patterns (Wong and Ghosh, 2002). The increase in dendritic complexity observed in the MeCP2 S421A cortical neurons suggests that phosphorylation of MeCP2 in response to activity might limit the initial phases of dendritic outgrowth, or could help to refine the pattern of dendritic arborization in response to synaptic signaling in subsequent phases of dendritic development.
Figure 2. Increased Dendritic Complexity Upon Loss of MeCP2 S421 Phosphorylation.
(A) Dissociated E16+21–22 DIV cortical neurons from MeCP2 S421A knock-in mice or wild-type littermates transfected with GFP to visualize dendritic morphology. Maximal intensity projections of a series of confocal planes for representative 25X images are shown.
(B) Sholl analysis measuring the number of dendritic branch points at 10 μm radial increments from the cell soma reveal an increase in dendritic branching in the MeCP2 S421A cells (p < 0.05, two-way repeated measures ANOVA) due to increased branch points 60–80 μm from the cell soma (p < 0.01, Bonferroni posttests). Wild-type (n=26) and MeCP2 S421A knock-in (n=33) neurons from two independent experiments.
(C) Representative images of single layer V pyramidal neurons expressing GFP under from the Thy1 promoter in 4-week-old forebrains of GFP-M transgenic mice. Tracings are shown of maximal intensity projections of a series of 10X confocal planes taken from coronal cortical sections.
(D) Sholl analysis of dendritic branch points at 10 μm radial increments from the cell soma reveals an increase in dendritic branching of MeCP2 S421A layer V pyramidal neuron apical dendrites 530 (p < 0.05), 540 (p < 0.01), 550 (p < 0.01) and 560 μm (p < 0.05) from the cell soma (two-way repeated measures ANOVA with Bonferroni posttests). Wild-type (n=29) and MeCP2 S421A knock-in (n=16) neurons from three independent littermate pairs.
(E) The maximum number of dendritic intersections ≥ 400 μm from the cell soma observed by Sholl analysis of each GFP-M+ layer V pyramidal cell in MeCP2 WT and S421A knock-in mice. The increase in maximum number of intersections in the MeCP2 S421A knock-in cells compared to WT indicates that the MeCP2 S421 knock-in apical tuft is more complex regardless of the exact distance of the apical tuft from the cell soma. p < 0.01 by Student’s t-test. All data shown in Figure 2 are mean ± SEM.
Pyramidal neurons are found primarily in forebrain structures, and their distinct patterns of dendritic branching determine the response of the cell to synaptic inputs (Spruston, 2008). To investigate whether MeCP2 S421 phosphorylation contributes to dendritic patterning in the cortex in vivo, we crossed S421A knock-in mice to the GFP-M transgenic line which expresses enhanced green fluorescent protein in a sparse subpopulation of neurons throughout the brain (Feng et al., 2000), facilitating morphological analysis of individual neurons. We examined GFP-positive cortical layer V pyramidal cells in these experiments because previous studies have shown that the disruption of MeCP2 function results in defects in dendritic growth of layer V neurons, and also disrupts the synaptic connectivity of layer V neurons within cortical circuits (Armstrong, 2002; Dani, 2005; Dani and Nelson, 2009).
Sholl analysis of layer V pyramidal cells from MeCP2 S421A mice and their wild-type littermates revealed a striking increase in the complexity of MeCP2 S421A dendrites specifically within the distal apical tuft located between 530 and 560 μm from the cell soma, corresponding to the region of the apical tuft that extends into layer I (Figure 2C–E). Importantly, sensory experience is required for the formation and maintenance of the dendritic spines of these apical tuft dendrites (Holtmaat and Svoboda, 2009), and the regulation of dendritic complexity by MeCP2 S421 phosphorylation may represent a related mechanism of experience-dependent neuronal development. In contrast to apical tuft dendrites, the complexity of basal and proximal apical dendrites did not differ significantly between the MeCP2 S421A and wild-type cells. The fact that in cortical circuits layer V pyramidal cells receive distinct types of synaptic input on their basal, proximal apical, and distal apical dendrites, together with our finding that MeCP2 S421 phosphorylation is selectively required for the patterning of the distal apical dendrites, suggests that MeCP2 phosphorylation at S421 may be critical for specific aspects of activity-dependent circuit development. Moreover, disruptions in this activity-dependent process when MECP2 is mutated may contribute to features of RTT.
Loss of MeCP2 S421 Phosphorylation Shifts Excitation-Inhibition Balance
Given the importance of neuronal patterning for synaptic connectivity, we reasoned that the increase in dendritic complexity observed in the MeCP2 S421A cortex might reflect a role for activity-dependent MeCP2 phosphorylation in cortical circuit development. While it has been proposed that MeCP2 regulates experience-dependent aspects of synaptic maturation, the specific importance of activity-dependent regulation of MeCP2 in this context has not been demonstrated. To assess the effect of loss of MeCP2 S421 phosphorylation on synaptic development, we prepared acute slices containing primary visual cortex (V1) from the brains of postnatal day 16–17 MeCP2 S421A mice and their wild-type littermates, and obtained whole-cell patch-clamp recordings from layer II/III pyramidal neurons. Disruption of MeCP2 expression in the cortex has indicated a general requirement for MeCP2 in cortical neuron synaptic function (Guy et al., 2010). We analyzed pyramidal cells in cortical layer II/III of postnatal visual cortex because of the well-defined role of neuronal activity in modulating the function of these cells (Maffei and Turrigiano, 2008; Trachtenberg et al., 2000).
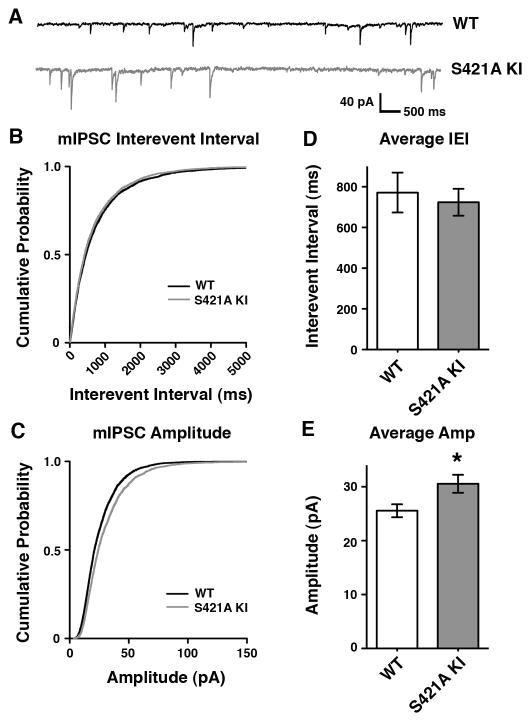
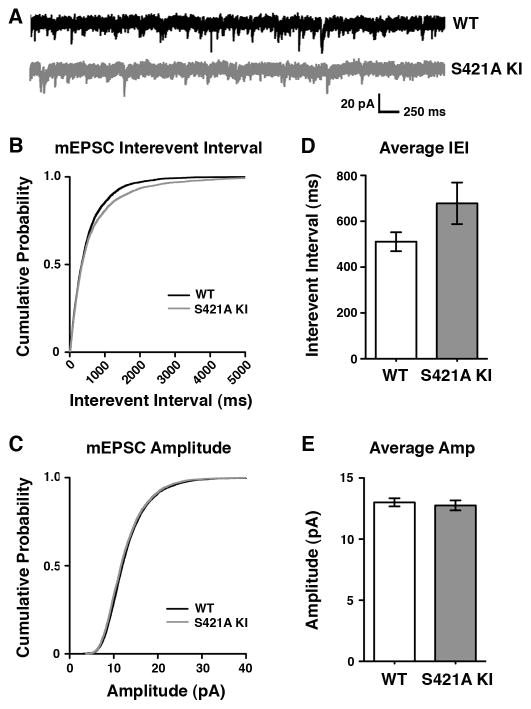
We analyzed pharmacologically isolated V1 layer II/III spontaneous miniature inhibitory postsynaptic currents (mIPSCs). We saw no effect on the frequency of mIPSCs obtained from these whole-cell patch-clamp recordings when wild-type and MeCP2 S421A mice were compared (Figure 3B and D). In contrast, the amplitude of mIPSCs recorded from MeCP2 S421A neurons in acute cortical slice were significantly increased relative to neurons from wild-type samples (Figure 3C and E). The increase in mIPSC amplitude observed in the MeCP2 S421A brain suggests that the activity-dependent phosphorylation of MeCP2 functions in the regulation of inhibitory synaptic strength, and might reflect an increase in vesicle neurotransmitter content or postsynaptic GABA receptor number. Analysis of pharmacologically isolated spontaneous miniature excitatory postsynaptic currents (mEPSCs) suggested a trend towards a decrease in the frequency of mEPSC events in MeCP2 S421A knock-in neurons compared to wild-type neurons, although this change was not statistically significant (Figure 4).
Figure 3. Loss of Activity-Dependent MeCP2 S421 Phosphorylation Regulates Development of Inhibitory Synaptic Transmission in vivo.
(A) Representative traces of mIPSCs recorded from layer II/II V1 pyramidal neurons in acute cortical slices from P16–17 MeCP2 S421A knock-in mice or their wild-type littermates.
(B–C) Cumulative probability distribution of mIPSC interevent intervals (B) and amplitudes (C) recorded from MeCP2 S421A or wild-type littermates.
(D–E) Average interevent interval (D) and amplitude (E) of mIPSCs recorded from wild-type or MeCP2 S421A neurons. Data are mean ± SEM. The difference in amplitude is statistically significant, p < 0.05 by Student’s t-test.
Data shown represent 250 random events drawn from each of the wild-type (n=19) or MeCP2 S421A knock-in (n=21) cells analyzed, recorded from mice from 6 independent litters.
Figure 4. Analysis of mEPSCs in the MeCP2 S421A Knock-In Mouse.
(A) Representative traces of mEPSCs recorded from layer II/II V1 pyramidal neurons in P16–17 MeCP2 S421A knock-in mice or their wild-type littermates.
(B–C) Cumulative probability distributions of mEPSC interevent intervals (B) or amplitudes (C) recorded from MeCP2 S421A or wild-type littermates.
(D–E) Average interevent interval (D) and ampltitude (E) of mEPSCs recorded from MeCP2 S421A or wild-type neurons. Data are mean ± SEM, p = 0.1 by Student’s t-test.
Data shown represent 250 random events drawn from each of the wild-type (n=23) or MeCP2 S421A knock-in (n=25) cells analyzed, recorded from 6 pairs of littermates.
Together these findings suggest that activity-dependent MeCP2 S421 phosphorylation is required for the proper development of synaptic connections within cortical circuits. Notably, the overall shift in excitation-inhibition balance in the MeCP2 S421A knock-in brain is similar in both direction and magnitude to that described in the MeCP2 knockout animal (Dani, 2005). The observed shift in the balance of synaptic inputs onto pyramidal cells in favor of inhibition in the MeCP2 S421A knock-in cortex suggests that loss of the activity-dependent phosphorylation of MeCP2 S421 may contribute to the synaptic defects that have been observed in other mouse models of RTT. Moreover the finding that S421 phosphorylation of MeCP2 is important for the development of cortical inhibitory synapses is consistent with the recent appreciation for the importance of activity-dependent programs of gene expression in regulating the development of inhibition (Hong et al., 2008; Lin et al., 2008).
MeCP2 S421A Mutation Results in Abnormal Behavioral Responses to Novelty
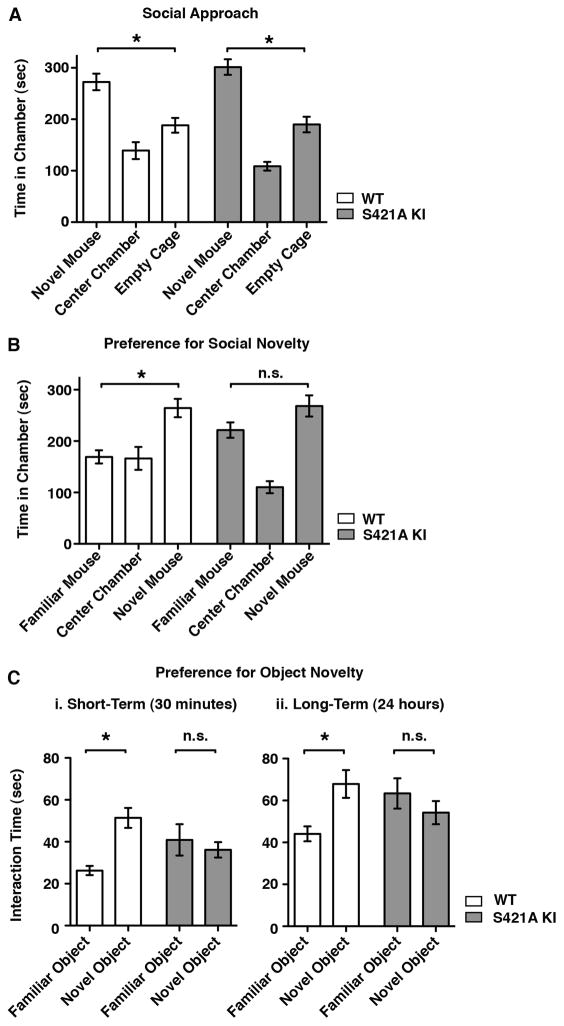
The alterations in cortical dendritic morphology and synaptic function observed in MeCP2 S421A mice support the hypothesis that activity-dependent regulation of MeCP2 in neurons is critical for normal brain development. Disruptions in brain development such as those seen in the MeCP2 S421A mice can have a profound impact on adaptive responses of the nervous system throughout life, suggesting that the MeCP2 S421A mutation might result in abnormal behavior in adult MeCP2 S421A mice. We found that MeCP2 S421A mice are visually indistinguishable from their wild-type littermates and show no major abnormalities in motor activity levels or function (Figure S2). This made it possible for us to assess whether MeCP2 S421A mice might be abnormal in their responses to input from their environment. Given the importance of MeCP2 in humans in the development of neural circuits that underlie social functions and adaptability, we analyzed the behavior of MeCP2 S421A mice using an assay that was developed to assess sociability and the preference for social novelty in mice (Moy et al., 2004).
MeCP2 S421A knock-in mice or their wild-type littermates were placed in a three-chambered arena, and the behavior of the mice in this environment was monitored. A novel mouse that the test subject had never before encountered was placed within a small wire cage in one of the side-chambers of the arena. The test subject chose whether or not to enter the chamber containing the novel mouse, and the amount of time spent in each chamber of the arena was recorded. Wild-type mice are social animals and choose to spend more time in the chamber with another mouse (Figure 5A). This social interaction behavior was unaffected in MeCP2 S421A mice, demonstrating their ability to recognize other mice and their appropriate interest in their physical and social environment. Subsequently, a second mouse that the test subject had never before encountered was placed within a small wire cage in the side-chamber opposite to the first, now familiar mouse. The wild-type test subjects spent the largest proportion of their time in the chamber containing the second, novel mouse and less time with the familiar choice (Figure 5B). By contrast, the MeCP2 S421A mice spent as much time with the familiar mouse as with the novel mouse.
Figure 5. Defects in Behavioral Response to Novel Experience in MeCP2 S421A Knock-In Mice.
(A) Sociability is not affected by loss of MeCP2 S421 phosphorylation. Behavior of wild-type (n = 24) and MeCP2 S421A knock-in (n=21) mice in a three-chamber apparatus with a single novel mouse placed in a small wire cage on one of the side chambers. Data shown are time spent in each chamber over the 10 minute trial period. Wild-type and S421A mice spent significantly more time in the chamber with the novel mouse (WT p < 0.001, S421A p < 0.01). In this assay wild-type and MeCP2 S421A mice were statistically indistinguishable.
(B) MeCP2 S421A mice do not lose interest in familiar mice. Behavior of wild-type (n = 24) and MeCP2 S421A knock-in (n=21) mice in a three-chamber apparatus with a familiar mouse placed in one of the side chambers and a novel mouse placed in the opposite side chamber. Time spent in each chamber over the 10 minute trial period is shown. Wild-type mice spent significantly more time with the novel mouse than with the familiar mouse or alone (p < 0.01). The amount of time spend with each test mouse did not differ significantly for MeCP2 S421A knock-in mice.
(C) MeCP2 S421A mice do not distinguish between familiar and novel objects. Behavior of wild-type (n = 9) and MeCP2 S421A (n = 9) mice when placed in an arena with a familiar inanimate object and a novel inanimate object placed at opposite ends. In the short-term assay (i), the familiar object was first introduced 30 minutes prior to the trial. In the long-term assay (ii), the familiar object was first introduced 24 hours prior to the trial. Wild-type mice spend significantly more time interacting with the novel object than with the familiar object in both assays (i, p < 0.01; ii, p < 0.05), while MeCP2 S421A knock-in mice do not. Total time spent exploring each object over a 10 minute trial period is shown for each assay.
Data for all plots are mean ± SEM, p-values from one-way ANOVA with Bonferroni multiple comparison correction.
Because the MeCP2 S421A mice show appropriate interest in novel mice, it is unlikely that the increased time spent with familiar mice is due to a general deficit in social recognition upon loss of MeCP2 S421 phosphorylation. Likewise, the MeCP2 S421A mice show no aversion to spending time alone and appear normal in tests of anxious behavior (Figure S2). Instead the increased interest in the familiar mouse suggests that the MeCP2 S421A mutants cannot distinguish between familiar and novel mice. This lack of discrimination between novel and familar stimuli exhibited by the MeCP2 S421A mice was not limited to social behavior; when presented with both novel and familiar inanimate objects, wild-type mice showed a behavioral preference for a novel object, while the MeCP2 S421A mice spent equal amounts of time investigating both familiar and novel objects (Figure 5C). This difference was evident at 30 minutes after the initial exposure to the familiar object, and persisted even after 24 hours had passed.
Taken together, these findings support the conclusion that neuronal activity-dependent phosphorylation of MeCP2 at S421 is necessary to allow an animal to process novel experience and respond appropriately to previously encountered objects or animals. This defect cannot be attributed to an absence of all learning and memory in these mice, as the performance of the MeCP2 S421A mice in spatial learning and memory tests is indistinguishable from wild-type (Figure S3). Instead the specific defect observed in the MeCP2 S421A mice suggests that the activity-dependent phosphorylation of MeCP2 S421 contributes to aspects of cognitive function underlying behavioral flexibility, and that the disruption of this aspect of MeCP2 regulation in RTT may be a factor in cognitive impairments observed in affected individuals.
MeCP2 Binds to the Neuronal Genome in a Histone-Like Fashion
The abnormalities we observe in the MeCP2 S421A knock-in animals demonstrate that this activity-dependent phosphorylation event is required for proper formation of the nervous system. We considered how the phosphorylation of S421 might modulate the molecular function of MeCP2 during neuronal development. Two distinct mechanisms have been proposed to explain how MeCP2 functions when bound to DNA. One model suggests that MeCP2 acts as a transcriptional repressor, binding at the promoters of specific target loci to prevent gene activation in particular contexts. For example, semi-quantitative chromatin immunoprecipitation (ChIP) experiments suggested that in neurons MeCP2 is bound specifically to the promoter of Bdnf to repress the expression of this activity-dependent gene. In response to neuronal activation the phosphorylation of MeCP2 at S421 was proposed to decrease MeCP2 binding to the Bdnf promoter, relieving repression and thus permitting Bdnf transcription (Chen et al., 2003; Martinowich, 2003). Subsequent studies have shown that MeCP2 binds near both active and repressed genes (Chahrour et al., 2008; Wu et al., 2010; Yasui et al., 2007), questioning whether binding of MeCP2 at specific loci is sufficient to repress transcription. Most recently, MeCP2 ChIP-Seq experiments demonstrated that MeCP2 binds broadly throughout the genome (Skene et al., 2010), suggesting that MeCP2 functions more like a histone protein than a sequence-specific transcription factor. Thus it is possible that MeCP2 regulates chromatin state globally rather than controlling the level of gene transcription at specific loci.
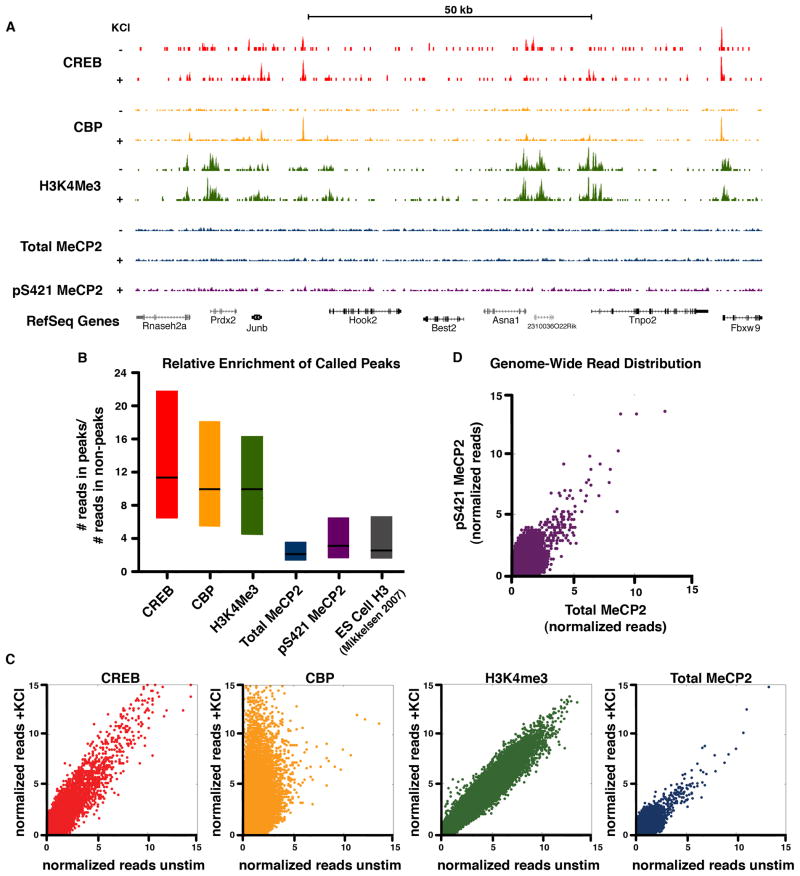
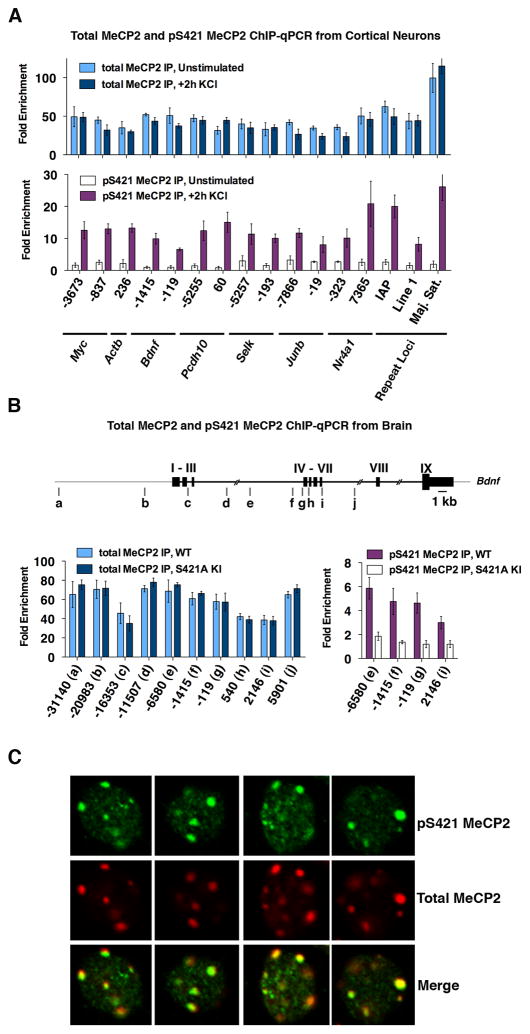
To assess what effect S421 phosphorylation has on MeCP2 function, we determined the sites of MeCP2 binding across the genome in untreated and membrane-depolarized neurons using MeCP2 ChIP followed by high-throughput sequencing (ChIP-Seq). We used antibodies raised against the C-terminus of MeCP2 that recognizes the protein regardless of its phosphorylation state. We confirmed the specificity of the anti-MeCP2 antibodies for ChIP from mouse brain and cultured cortical neurons using quantitative PCR across the Myc locus, a region shown to be bound by MeCP2 (Skene et al., 2010) (Figure S4A). We then performed ChIP-Seq from cultured neurons that were either left unstimulated or membrane depolarized with high extracellular KCl for 2 hours. Mapping of the MeCP2 ChIP-Seq reads to the genome revealed a broad distribution of MeCP2 in both unstimulated and membrane depolarized neurons (Figure 6A). MeCP2 ChIP-qPCR confirmed that the distribution of reads obtained by ChIP-Seq represents broad distribution of MeCP2 across the genome rather than failure of the ChIP to enrich for MeCP2-bound DNA: we observed at least 20-fold enrichment above peptide-blocked control ChIP at all sites tested, including regions surrounding activity-regulated genes, constitutively active loci, and repetitive genomic elements (Figure 7A).
Figure 6. ChIP-Seq analysis reveals a histone-like binding profile for total MeCP2 and global phosphorylation of MeCP at S421 upon neuronal stimulation.
(A) ChIP-Seq profiles for total and pS421 MeCP2 as well as CREB, CBP and H3K4me3 (Kim et al., 2010) across a representative genomic region. ChIP samples were isolated from E16 + 7 DIV cortical cultures, unstimulated (‘−‘) or stimulated for 2 hr with 55mM KCl (‘+’). The location of RefSeq genes, including the activity-regulated immediate-early gene Junb, are indicated.
(B) Box plots (1st quartile, median, and 3rd quartile) displaying the ratio of the number of reads found at called peaks versus non-peak control regions are shown for each ChIP-seq data set (2hr KCl stimulated). Enrichment for pan-histone H3 ChIP-Seq peaks from embryonic stem cells is shown for comparison (H3ES, Mikkelsen et al., 2008).
(C) Scatter plots depict normalized ChIP-Seq reads from E16 + 7 DIV cortical cultures for 1000 bp windows tiled across the genome before and after stimulation (2hr KCl). Data points on the diagonal indicate no substantial change in ChIP signal upon stimulation. Points located off the diagonal, as seen for CBP, indicates dynamic change in response to neuronal activity.
(D) Scatter plot depicts normalized ChIP-Seq reads of pS421 MeCP2 versus total MeCP2 from stimulated dissociated cortical cultures (E16 + 7 DIV, 2 hr KCl) for 1000 bp windows tiled across the genome. Distribution along the diagonal indicates that the profile of pS421 reads tracks with total MeCP2 reads across the genome.
Figure 7. ChIP-qPCR and immunohistochemistry confirm broad binding of total MeCP2 and global MeCP2 S421 phosphorylation in vitro and in vivo.
(A) ChIP-qPCR analysis of total MeCP2 (top panel) and pS421 MeCP2 (bottom panel) in cultured cortical neurons (E16 + 7 DIV) that were either membrane depolarized for 2 hours (55mM KCl) or left unstimulated.
(B) Total MeCP2 (left) and pS421 MeCP2 (right) ChIP-qPCR from the forebrains of 7-week-old MeCP2 S421A mice or their wild-type littermates. The location of PCR amplicons across the Bdnf locus are shown in diagram as a-j. Nucleotide position is given relative to start of the activity-dependent Bdnf promoter IV.
All ChIP data is presented as fold enrichment above a negative control ChIP performed in parallel on each sample using the same antiserum that had been preincubated with the peptide antigen to which it was raised. Data are mean ± SEM from three independent experiments.
(C) Immunocytochemistry of nuclei from wild-type E16 + 10 DIV mouse cortical neurons following 30 min membrane depolarization with 55 mM KCl. Antibodies used are specific for total MeCP2 or pS421 MeCP2 (see also Figure S4). Representative 63X images from single confocal planes are shown.
The broad distribution of MeCP2 that we detect is similar to that previously reported (Skene et al., 2010). Notably, the previous MeCP2 ChIP analysis was carried out using brain extracts and an antibody that recognizes an N-terminal region of MeCP2. In the present study we have used cultured neurons and an anti-C-terminal-MeCP2 antibody. The fact that similar results are obtained using two different anti-MeCP2 antibodies provides support for the conclusion that MeCP2 binds broadly across the neuronal genome.
The relatively uniform distribution of MeCP2 ChIP-Seq reads that we observe is inconsistent with the idea that MeCP2 binds at discrete sites within the regulatory regions of target genes. This point is illustrated by comparison of the pattern of MeCP2 binding to that of canonical transcription factors. For example, CREB is a sequence-specific DNA-binding factor critical for activity-dependent gene regulation in neurons that has been suggested to associate with MeCP2 (Chahrour et al., 2008). Previous CREB ChIP-Seq analysis performed in neuronal cultures identical to those used for our MeCP2 ChIP-Seq (Kim et al., 2010) demonstrates that CREB binds to the genome at discrete sites (Figure 6A). Peaks of CREB binding are defined by a peak detection algorithm that identifies regions with high enrichment of reads relative to the average genomic distribution (Figure 6B). In contrast, genome-wide peak detection analysis of total MeCP2 ChIP reads identified modest fluctuations in protein binding, and the read enrichment for these regions was substantially lower than that of the CREB peaks (Figure 6B and data not shown).
Instead the read distribution of MeCP2 across the genome is quite similar to histone H3 ChIP-Seq data obtained from mouse embryonic stem cells (Mikkelsen et al., 2008), suggesting that MeCP2 may function more like a histone than a classical transcriptional repressor. To compare the profile of H3 to MeCP2, we performed genome-wide peak detection analysis for this histone H3 ChIP-Seq and, as for MeCP2, were able to identify only modest fluctuations in protein binding with relatively low read enrichment within these identified regions (Figure 6B). The similarities between the binding profiles of MeCP2 and histone H3 support the hypothesis that MeCP2 functions as a core component of neuronal chromatin.
MeCP2 Binding Profiles are Maintained During Neuronal Activation
It has been suggested that neuronal activation and subsequent phosphorylation of MeCP2 reduces the affinity of MeCP2 for DNA, providing a mechanism through which extracellular stimuli could regulate neuronal chromatin. Local alterations in the binding of MeCP2 at particular genes in response to neuronal stimuli could allow this histone-like factor to affect the transcription of individual target genes. To test this hypothesis, we examined MeCP2 binding profiles in neurons before and after membrane depolarization. Genome-wide comparisons by MeCP2 ChIP-Seq revealed no evidence of robust changes in MeCP2 binding between the two conditions (Figure 6C). Moreover, computational searches for regions of significantly increased or reduced binding failed to detect reproducible changes in the relative enrichment of MeCP2 upon neuronal activation. In contrast, analysis of ChIP-Seq data for the transcriptional coactivator CBP in the identical neuronal culture and stimulation paradigm (Kim et al., 2010) reveals a dramatic increase in the binding of CBP across the genome in response to membrane depolarization, demonstrating our ability to detect dynamic changes in factor binding (Figure 6C).
We note that the widespread distribution of MeCP2 ChIP-Seq reads across the genome limits our power to detect a loss of MeCP2 binding occurring over small regions due to low read coverage. To address this, we analyzed MeCP2 binding at multiple loci before and after membrane depolarization in neurons using more targeted and sensitive ChIP-qPCR. We failed to detect any significant changes in MeCP2 binding across the promoters of multiple activity-dependent genes in neuronal cultures (Figure 7A), or in the brains of wild-type and MeCP2 S421A mice (Figures 7B and S4). In addition, we detected no activity-induced changes in MeCP2 ChIP signal at a number of constitutively expressed genes and repetitive loci (Figure 7A and S4). These data indicate that in this neuronal stimulation paradigm, where ~10–30% of the MeCP2 molecules become newly phosphorylated at S421 (Figure 1B and data not shown), there is no evidence of dynamic changes in MeCP2 binding. While it remains possible that phosphorylation of additional sites or distinct stimulation conditions lead to dissociation of MeCP2 from the genome as reported previously at the Bdnf locus (Chen et al., 2003; Martinowich, 2003), we conclude that phosphorylation of MeCP2 S421 alone is not sufficient to release MeCP2 from DNA. It is possible that the previous reports demonstrating decreased MeCP2 binding to DNA upon membrane depolarization reflect that fact that the ChIP assays used at the time were semi-quantitative and therefore more subject to error.
Activity-Dependent Transcriptional Programs are not Affected in S421A Mutants
While our MeCP2 ChIP analysis suggests that neuronal activity does not induce changes in the binding of MeCP2 to DNA, it remained possible that the phosphorylation of MeCP2 bound to the promoters of activity-regulated genes might regulate activity-dependent gene transcription. To examine this possibility, we compared the level of activity-dependent Bdnf expression in dissociated primary cortical cultures from the brains of MeCP2 S421A mice and their wild-type littermates by RT-qPCR. Given previous studies showing that when MeCP2 is over-expressed in neurons the phosphorylation of MeCP2 at S421 affects Bdnf transcription (Zhou et al., 2006), we were surprised to find that the extent and time course of Bdnf induction upon membrane depolarization was not significantly different between wild-type and MeCP2 S421A neurons (Figure S5, and data not shown). Likewise, the kinetics of induction of other activity-regulated genes, such as c-fos, were similar in wild-type and MeCP2 S421A neurons.
Given these results, we broadened our approach and used Affymetrix GeneChip Mouse Expression Set 430 2.0 microarrays to assess whether loss of MeCP2 S421 phosphorylation affected global profiles of activity-dependent gene expression. Cortical neurons isolated from the brains of embryonic MeCP2 S421A mice and their wild-type littermates were grown for 7 DIV then depolarized with KCl for 1 or 6 hours. While we detected more than 300 genes whose expression levels changed at least three-fold in response to extracellular KCl, we found no significant differences in the number, extent, or time course of induction of activity-dependent mRNAs between the wild-type and MeCP2 S421A cells (Figure S5). We also used Mouse Gene 1.0 ST microarrays to assess mRNA levels in the visual cortex of postnatal day 16–17 wild-type and MeCP2 S421A knock-in brains, a timepoint and brain region where MeCP2 is phosphorylated at S421 (Figure S6). An analysis of transcript levels using either individual probes within specific gene regions or groups of probes across individual genes revealed no detectable mRNA dysregulation in the developing MeCP2 S421A mouse brain (data not shown). It remains possible that S421 phosphorylation has more subtle effects on the magnitude or timing of activity-dependent gene transcription. Alternatively, MeCP2 S421 phosphorylation may control aspects of transcription (e.g. initiation, elongation, termination, or the rate of transcription) or chromatin remodeling (e.g. histone modification and compaction or DNA methylation) that are not detected by measuring steady-state mRNA levels. It is also possible that other mechanisms of gene regulation largely compensate in the absence of MeCP2 S421 phosphorylation.
Phosphorylation of MeCP2 S421 Occurs Globally in Response to Neuronal Stimulation
To further investigate how S421 phosphorylation might affect MeCP2 function, we employed ChIP-Seq to examine where across the genome MeCP2 becomes newly phosphorylated at S421 in response to neuronal activity. We hypothesized that if pS421 MeCP2 controls a specific step in the process of activity-dependent gene transcription, then this phosphorylation event would be expected to occur at select regions of the genome (e.g. the promoters, enhancers, or exons of activity-dependent genes).
We first sought to establish the utility of our anti-pS421 MeCP2 specific antiserum for ChIP analysis (Figures 1A and S4B). We immunoprecipitated pS421 MeCP2 from unstimulated or KCl-depolarized neurons, and from the brains of wild-type or MeCP2 S421A knock-in mice. qPCR analysis of several loci demonstrated that the anti-pS421 MeCP2 antibody specifically recognizes the phosphorylated form of MeCP2 in ChIP assays: pS421 MeCP2 was found to be bound at all sites tested in membrane-depolarized neurons but not in unstimulated neurons where the level of MeCP2 S421 phosphorylation is quite low (Figure 7A), and was enriched in wild-type brain compared to MeCP2 S421A brain (Figure 7B). Moreover, the pS421 MeCP2 ChIP signal was competed away if the anti-pS421 MeCP2 antiserum was pre-incubated with the phospho-peptide antigen used to generate the antibody (Figure S4C).
To determine where along the genome MeCP2 S421 becomes phosphorylated in response to neuronal activation, we performed high-throughput sequencing of pS421 MeCP2 ChIP DNA isolated from cultured cortical neurons treated with 55mM KCl for 2 hours. Mapping of the sequenced pS421-MeCP2 ChIP reads to the genome revealed a strikingly uniform distribution of pS421 MeCP2 with little evidence of discrete peaks (Figure 6A), indicating that upon membrane depolarization MeCP2 bound broadly across the genome is phosphorylated at S421.
We considered the possibility that the genomic distribution of pS421 MeCP2 might be similar to that previously reported for specific histone modifications. Although histone modifications can be broadly distributed across the genome, it is possible to identify genomic elements that bear a particular mark while other regions of the genome are devoid of the histone modification. For example, while histone H3 lysine 4 trimethylation (H3K4me3) can span ~500 base pair to 2 kilobase surrounding promoters, there are genomic regions where this mark is absent (Zhou et al., 2011). Other marks, such as H3 S10 phosphorylation occur throughout the genome (Nowak and Corces, 2004). Importantly, the presence of these histone marks can be informative. For example, the H3K4me3 mark occurs at expressed genes, while H3S10 phophorylation is found across the genome as a hallmark of mitosis.
To scan for regions of the genome that are enriched for pS421 MeCP2 we searched for peaks across the genome, changing the parameters of the peak detection algorithm to detect peaks of different length scales. The loci identified by this approach revealed only very modest increases in sequencing reads relative to non-peak regions of the genome, supporting the conclusion that in membrane depolarized neurons pS421 MeCP2 is widespread across the genome (Figure 6B).
Finally, we considered the possibility that within the ubiquitous distribution of pS421 MeCP2 across the genome there might still be regions of relative enrichment. Since the pS421 MeCP2 ChIP-Seq analysis revealed some modest peaks of MeCP2 binding, we used ChIP-qPCR to ask if these putative pS421 MeCP2 peaks could be validated. However, an analysis of nine candidate peaks revealed that none displayed greater than a two-fold increase in signal above flanking, non-peak control regions. Importantly, all peak and non-peak regions tested displayed robust pS421 MeCP2 ChIP signal from depolarized neurons relative to pS421 MeCP2 ChIP signal from unstimulated neurons (Figure S4F), supporting the conclusion that upon membrane depolarization MeCP2 located throughout the genome becomes newly phosphorylated at S421. Indeed, in membrane depolarized neurons, genome-wide comparison of the pS421 and total MeCP2 ChIP-Seq reads shows that pS421 MeCP2 tracks well with total MeCP2 (Figure 6D). The widespread phosphorylation of MeCP2 is also evident in the brain where pS421 MeCP2 ChIP-qPCR across multiple loci shows a strong correlation to total MeCP2 ChIP-qPCR and significant enrichment above parallel ChIP experiments performed with MeCP2 S421A brain (Figures 7B and S4). Finally, immunostaining with the anti-pS421 MeCP2 antibody revealed that in wild-type neurons pS421 MeCP2 co-localizes with total MeCP2 throughout the nucleus (Figure 7C). Taken together, these observations indicate that in response to membrane depolarization in neurons, phosphorylation of S421 is evenly distributed across MeCP2 molecules bound throughout the genome. Phosphorylation therefore occurs at active and repressed promoters; intronic, exonic, and intergenic sequences; and repetitive regions and transposon sequences. Thus, rather than serving as a locus-specific mechanism for regulating the expression of particular mRNA transcripts, MeCP2 S421 phosphorylation appears to facilitate a global chromatin response to neuronal activation that likely underlies some aspect of chromatin remodeling that occurs in response to neuronal activity. The absence of this response in MeCP2 S421A mice may account for the dendritic, synaptic, and behavioral defects that we observe.
DISCUSSION
Intense investigation has focused on determining how mutations of MECP2 lead to RTT and related neurological disorders. The postnatal time course of RTT symptom onset together with the synaptic defects observed in Mecp2 mutant mice have led to the hypothesis that RTT is a disorder of experience-dependent synapse maturation. However, the devastating consequences of loss or over-expression of MeCP2 on cell and organismal health have made it difficult to assess whether defects in experience-dependent synaptic and cognitive development arise directly from, or are indirect consequences of, loss of MeCP2 function. Indeed, careful observation of individuals with RTT has suggested that different mutations in MECP2 can lead to distinct cognitive and clinical sequelae (Neul et al., 2008), suggesting that MeCP2 has a number of discrete roles in the development of the nervous system.
The discovery that experience induces the phosphorylation of MeCP2 at S421 in the brain revealed a mechanism by which neuronal activity might modulate MeCP2 function, and has provided a molecular handle to dissect the activity-dependent and -independent functions of MeCP2. In the present study we eliminated the neuronal activity-dependent phosphorylation of MeCP2 at S421 in vivo without otherwise affecting MeCP2 expression. By studying these MeCP2 S421A mice we find that MeCP2 S421 phosphorylation is required for the normal development of neuronal dendrites and inhibitory synapses in the cortex, demonstrating the importance of the activity-dependent regulation of MeCP2 for the establishment of appropriate connectivity in the nervous system. In addition, we find that loss of MeCP2 S421 phosphorylation results in defects in behavioral responses to novel versus familiar mice or objects, indicating that activity-dependent MeCP2 phosphorylation regulates aspects of cognitive function. Based on these findings, we propose that the disruption of MeCP2 phosphorylation at S421 contributes to the cognitive impairments observed in RTT and other MECP2-dependent disorders.
Compared to the effects of deleting Mecp2, mutating MeCP2 S421 to an alanine has a relatively mild effect on nervous system development. For example, the gross motor abnormalities, body weight dysregulation, seizures, and certain learning and memory defects observed in the MeCP2 knockout appear not to rely on the activity-dependent phosphorylation of MeCP2 at S421. This could suggest that aspects of MeCP2-regulated neuronal function rely on neuronal activity-independent development processes. Alternatively, it is possible that other stimulus-dependent MeCP2 modifications (D. Ebert and M. Greenberg, unpublished results) may function either singly or in combination to regulate MeCP2-dependent neuronal responses.
It has been proposed, based on mass spectrometry analysis (Tao et al., 2009), that phosphorylation of MeCP2 also occurs at serine 424 (S424). A recent study reports that the mutation of both MeCP2 S421 and S424 to alanines in mice results in alterations in hippocampal learning and synapse biology as well as changes in MeCP2 binding and dysregulation of a small number of candidate genes examined (Li et al., 2011). The phenotypes reported in these mice are similar to the phenotypes observed when MeCP2 is over-expressed in mice (Chao et al., 2007; Collins, 2004) raising the possibility that the mutation of S424 to alanine leads to enhanced MeCP2 expression or activity.
In an effort to determine if neuronal activity induces the phosphorylation of MeCP2 S424 we have generated anti-phospho-S424 MeCP2-specific antibodies, but we have been unable to detect increased phosphorylation of MeCP2 S424 in response to neural activity in vitro (KCl depolarized vs unstimulated cortical cultures,) or in vivo (kainate seized vs unseized brain) (D. Ebert and M. Greenberg, unpublished results). While it remains possible that MeCP2 S424 is phosphorylated constitutively or in response to other stimuli, we have restricted our analysis to the verified activity-dependent phosphorylation of MeCP2 at S421, allowing us to unambiguously relate the phenotypes we observe in MeCP2 S421A mice to activity-dependent MeCP2 phosphorylation.
Our observations using MeCP2 S421A mice reinforce the importance of in vivo models for studying the role of neuronal activity in nervous system development and function. Previous in vitro studies suggested a model in which, in the absence of neuronal activity, MeCP2 is bound to the promoters of activity-regulated genes such as Bdnf to repress their transcription (Chen et al., 2003; Martinowich, 2003; Zhou et al., 2006). Membrane depolarization-induced S421 phosphorylation was proposed to lead to reduced binding of MeCP2 at these activity-dependent promoters, relieving repression and allowing for gene activation. If this model were correct we would predict that neurons from MeCP2 S421A mice might demonstrate a defect in the induction of Bdnf or other activity-regulated genes. Instead, we found that the activity-dependent expression of Bdnf and other neuronal activity-dependent genes was not detectably dysregulated by loss of MeCP2 phosphorylation at S421, suggesting that this model of activity-dependent MeCP2 function must be revised.
To reassess the molecular function of MeCP2 and its regulation by neuronal activity, we turned to ChIP-Seq to examine the binding profile of MeCP2 genome-wide. Recent work from brain had suggested that MeCP2 binds broadly across the genome (Skene et al., 2010). By demonstrating that MeCP2 is highly enriched throughout the genome in both the brain and dissociated cortical cultures that contain very few glial cells, we exclude the possibility that the broad binding of MeCP2 observed in the brain arises as a result of heterogeneous contributions from neuronal and glial populations. Consistent with this recent study (Skene et al., 2010), the pattern of binding we detect in brain and neurons suggests that MeCP2 binds preferentially to methylated DNA (i.e. reduced binding at TSS sites, increased binding at repeat DNA). However, MeCP2 binding is not limited to methylated loci, as we note a high level of signal in MeCP2 ChIP assays from brain and cultured neurons at sites where DNA methylation is presumably very low (e.g. the TSS for the highly-expressed Myc gene), or devoid of CpG residues over long stretches. Interestingly, the ChIP profile of MeCP2 in E16 + 7 DIV cortical cultures is more flat than that found in the brains of 7-week-old mice (e.g. Figure S4A), suggesting that changes in DNA methylation or MeCP2 expression levels during nervous system development may lead to an increase in the dynamic range of the MeCP2 binding profile.
Taken together, our ChIP data allow us to conclude that MeCP2 is bound throughout the neuronal genome in a pattern similar to that of a histone protein. Several studies have demonstrated that MeCP2 binds to the linker DNA between nucleosomes in vitro similarly to linker histone H1 (Ghosh et al., 2010; Nan et al., 1997), and that in vivo histone H1 levels are up-regulated in the MeCP2 knockout brain (Skene et al., 2010). Our data is consistent with a model in which MeCP2 takes the place of H1 molecules throughout the neuronal genome, functioning on a global scale to modulate chromatin structure.
By examining genome-wide profiles of MeCP2 before and after neuronal stimulation, we have assessed the potential for dynamic regulation of MeCP2 binding by activity-dependent phosphorylation. Under the conditions used for these experiments, S421 phosphorylation is induced on a substantial fraction of MeCP2 molecules, yet we do not detect changes in the profile of MeCP2 binding across the genome. Because of the broad distribution of MeCP2, low read coverage limits our power to detect discrete regions where binding may be lost. However, using more sensitive ChIP-qPCR at multiple candidate activity-dependent loci we are unable to detect stimulus-dependent changes in binding. Consistent with this finding, seizure-induced increases in neuronal activity do not detectably alter MeCP2 binding at the activity-regulated promoter of Bdnf (H. Gabel, B. Kinde and M. Greenberg, unpublished observations). While it remains possible that a small number of discrete sites experience changes in binding or that there is a subtle change in global binding within the variability of our experiments, our data suggest that a stimulus capable of robustly inducing MeCP2 S421 phosphorylation is not sufficient to cause MeCP2 dissociation from the genome. Furthermore, because our stimuli induce the expression of Bdnf and other activity-regulated genes, dissociation of MeCP2 from the DNA is not strictly required for transcriptional induction of these genes. Instead it appears that neuronal activity induces the phosphorylation of MeCP2 molecules that remain bound to the genome, serving to modulate MeCP2 function in situ.
Given the histone-like binding of MeCP2 to the neuronal genome, we considered that the phosphorylation of MeCP2 S421 could function in a manner analogous to a histone modification. While studies of pan-histone genomic binding profiles have provided important information about chromatin structure, ChIP analysis of specific histone modifications has led to a rich understanding of the localization and dynamics of these modifications, providing insight into their function in the modulation of gene expression (Zhou et al., 2011). As a first step toward understanding where post-translational modifications of MeCP2 occur on the genome, we performed ChIP analysis using a specific pS421 MeCP2 antiserum. We demonstrate that the neuronal activity-induced phosphorylation of S421 is evenly distributed across MeCP2 molecules bound to the genome. We estimate the percentage of MeCP2 phosphorylated at S421 in response to neuronal stimulation (2 hr KCl depolarization) to be 10–30%. If one MeCP2 molecule is bound every two nucleosomes as demonstrated by (Skene et al., 2010), and phosphorylation is evenly distributed across MeCP2 molecules, then an independent phosphorylation event is occurring approximately every 900–3000 bp. Thus, pS421 MeCP2 is likely to be extremely common across the genome, and has the potential to affect chromatin at a genome-wide scale. These findings suggest that instead of regulating specific target genes, MeCP2 S421 phosphorylation likely plays a more global role in modulating the response of neuronal chromatin to activity.
While many histone modifications have been found in discrete loci, genome-wide phosphorylation of histone H3 (e.g. H3S10) and histone H1 are thought to facilitate mitotic chromosomal rearrangements in non-neuronal cells (Happel and Doenecke, 2009; Nowak and Corces, 2004). This precedent suggests that the global phosphorylation of MeCP2 may alter chromatin compaction states throughout the nucleus or facilitate nuclear reorganization events that have been reported to occur in response to neuronal activity (Wittmann et al., 2009). As a result, the phosphorylation of MeCP2 S421 may play a role in fine-tuning aspects of gene expression that are not readily detected with standard measures of gene expression. Alternatively, the global phosphorylation of MeCP2 S421 could collaborate with locus-specific modifications of MeCP2: S421 phosphorylation would facilitate transcriptional processes specifically gated by other stimulus-dependent modifications of MeCP2. Either of these models could explain why we observe phenotypes consistent with defects in experience-dependent neuronal development upon loss of MeCP2 S421 phosphorylation, but are unable to detect significant changes in the expression of individual genes.
Together with recent analysis (Skene et al., 2010), our data support a conceptual shift in the understanding of the function of MeCP2 in neurons. Instead of acting solely as a repressor to regulate gene expression through targeted, dynamic binding to chromatin, it may be more appropriate to consider MeCP2 as a constitutive component of neuronal chromatin. The idea that MeCP2 has many functions is consistent with the discovery of multiple, independently occurring phosphorylation events on MeCP2 (Huttlin et al., 2010; Tao et al., 2009; Zhou et al., 2006) (D. Ebert and M. Greenberg, unpublished observations) and the finding that total loss or over-expression of MeCP2 leads to subtle changes in the expression of thousands of genes rather than derepression of a discrete subset of target genes. Just as different histone modifications can correlate with independent and often opposing effects on gene expression, the different modifications of MeCP2 may have distinct influences on chromatin at sites where they occur. The experiments presented here demonstrate that it is possible to perform modification-specific ChIP analysis of MeCP2 to gain insight into its global role. Future studies employing this approach may indicate where additional phosphorylation events occur on the genome and provide insight into how they modulate MeCP2 function.
By revealing the functional role of MeCP2 S421 phosphorylation, the MeCP2 S421A mouse demonstrates the utility of an in vivo approach for testing hypotheses regarding activity-dependent regulation of MeCP2. Moreover, the phenotypes observed in the MeCP2 S421A mice provide in vivo evidence that the stimulus-dependent modification of a chromatin regulator is required for nervous system development and function. Additional knock-in mutations to disrupt other activity-dependent modifications of MeCP2 may provide useful models for further study of stimulus-dependent regulation of MeCP2 in vivo, and should yield insight into the role of the environment in regulating neuronal chromatin function. Finally, given the critical importance of MeCP2 for nervous system development, future experiments to understand activity-dependent MeCP2 regulation will not only serve to deepen our understanding of stimulus-dependent chromatin biology, but should also provide therapeutic insight into RTT and other neuropsychiatric disorders.
EXPERIMENTAL PROCEDURES
Generation of MeCP2 S421A Knock-In Mice
Targeting of the MeCP2 locus was carried out by homologous recombination (Figure S1) in ES cells. A detailed description of animal breeding and genotyping is provided in the supplemental experimental procedures.
Neuronal Cell Culture, Protein Expression, Immunocytochemistry and Immunohistochemistry Analysis
Cortical or hippocampal neurons were prepared from E16 mouse embryos as described by (Xia et al., 1996). Western blotting, immunocytochemistry and immunohistochemistry tissues was carried out using standard methods. Sholl analysis of dendritic morphology was performed using Neurolucida (MBF bioscience) software on images acquired by confocal microscopy. For details see supplemental experimental procedures.
Chromatin Immunoprecipitation Analysis
ChIP experiments were performed as previously described (Kim et al., 2010) with some modifications. Immunopreciptiation was performed using custom rabbit polyclonal antisera recognizing either total MeCP2 or phosphoserine 421 MeCP2 (Zhou et al. 2006). ChIP-Seq was performed using the SOLiD instrument (Life Technologies) as previously described (Kim et al., 2010), with the slight modifications. For a description of computational analysis and experimental details see supplemental experimental procedures.
Gene Expression Analysis
For gene expression in cultured neurons or brain tissue, total RNA was isolated and cDNA generated using standard methods. qPCR analysis was carried out using the StepOnePlus system (Life technologies Beverly, MA). Microarray analysis of cultured wild-type and S421A neurons RNAs samples was carried out using Affymetrix GeneChip Mouse Expression Set 430 2.0 microarrays arrays. Microarray analysis from visual cortex employed total RNA isolated from P17 mice and analyzed on Affymetrix Mouse Gene 1.0 ST microarrays. See supplemental experimental procedures for details.
Electrophysiology
Whole-cell voltage clamp recordings were measured from layer II/III pyramidal neurons in coronal sections (250 μm) containing the primary visual cortex of P16–P18 mice. For both amplitude and interevent interval, 250 events were randomly selected from each cell. See supplemental experimental procedures for details.
Behavioral Analysis
The assay of sociability and preference for social novelty were based on a well-characterized assay from the Crawley lab (Moy et al., 2004). For details of all behavioral analyses see supplemental experimental procedures.
Supplementary Material
Acknowledgments
We thank E. Griffith, P. Greer, S. Ross, N. Robinson and Greenberg Lab members for critical reading of the manuscript; C. Chen and B. Bloodgood, E. Hong, R. Rodriguiz, J. Dodart, H. Ho and the MRDDRC Gene Manipulation Core (M. Thompson, Y. Zhou and H. Ye) for experimental assistance and advice. This work was supported by NIH grant 1R01NS048276 to MEG and R01 DA022202 to AEW; SC was supported by the Medical Scientist Training Program, Ruth L. Kirschstein NIH Training Grant T32CA009361 and the Dept. of Neurobiology Grad. Fellowship Fund at HMS; HWG was supported by Damon Runyon Cancer Research Foundation Grant DRG-2048-10; DHE was supported by NIH grant K08MH90306 and by the Dupont-Warren Fellowship in the Dept. of Psychiatry at HMS; ZZ was supported by NIH grant K99 NS058391. MH was supported by grants NIH 1R21NS070250-01A1, DP2 OD006461-01, and NSF 0954570.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armstrong DD. Neuropathology of Rett syndrome. Mental Retardation and Developmental Disabilities Research Reviews. 2002;8:72–76. doi: 10.1002/mrdd.10027. [DOI] [PubMed] [Google Scholar]
- Ballas N, Lioy DT, Grunseich C, Mandel G. Non cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nature Neuroscience. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong STC, Qin J, Zoghbi HY. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi H. The Story of Rett Syndrome: From Clinic to Neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chao HT, Zoghbi HY, Rosenmund C. MeCP2 Controls Excitatory Synaptic Strength by Regulating Glutamatergic Synapse Number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Collins AL. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Human Molecular Genetics. 2004;13:2679–2689. doi: 10.1093/hmg/ddh282. [DOI] [PubMed] [Google Scholar]
- Dani VS. Reduced cortical activity due to a shift in the balance between excitation and inhibition in a mouse model of Rett Syndrome. Proceedings of the National Academy of Sciences. 2005;102:12560–12565. doi: 10.1073/pnas.0506071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani VS, Nelson SB. Intact Long-Term Potentiation but Reduced Connectivity between Neocortical Layer 5 Pyramidal Neurons in a Mouse Model of Rett Syndrome. Journal of Neuroscience. 2009;29:11263–11270. doi: 10.1523/JNEUROSCI.1019-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nature Neuroscience. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, Lichtman JW, Sanes JR. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Ghosh RP, Horowitz-Scherer RA, Nikitina T, Shlyakhtenko LS, Woodcock CL. MeCP2 Binds Cooperatively to Its Substrate and Competes with Histone H1 for Chromatin Binding Sites. Molecular and Cellular Biology. 2010;30:4656–4670. doi: 10.1128/MCB.00379-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Cheval H, Selfridge J, Bird A. The Role of MeCP2 in the Brain. Annu Rev Cell Dev Biol. 2010 doi: 10.1146/annurev-cellbio-092910-154121. [DOI] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Holtmaat A, Svoboda K. Experience-dependent structural synaptic plasticity in the mammalian brain. Nat Rev Neurosci. 2009;10:647–658. doi: 10.1038/nrn2699. [DOI] [PubMed] [Google Scholar]
- Hong EJ, McCord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Li H, Zhong X, Chau KF, Williams EC, Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa I, Swanberg S, Harvey D, LaSalle JM, Jin LW. Rett Syndrome Astrocytes Are Abnormal and Spread MeCP2 Deficiency through Gap Junctions. Journal of Neuroscience. 2009;29:5051–5061. doi: 10.1523/JNEUROSCI.0324-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffei A, Turrigiano GG. Multiple modes of network homeostasis in visual cortical layer 2/3. J Neurosci. 2008;28:4377–4384. doi: 10.1523/JNEUROSCI.5298-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, Piven J, Crawley JN. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmuhl Y, Fischer D, Holsboer F, Wotjak CT, Almeida OF, Spengler D. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Nan X, Tate P, Li E, Bird A. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. doi: 10.1128/mcb.16.1.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, Zoghbi H, Percy A, Glaze DG. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 2004;20:214–220. doi: 10.1016/j.tig.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruston N. Pyramidal neurons: dendritic structure and synaptic integration. Nature Reviews Neuroscience. 2008;9:206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci U S A. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg JT, Trepel C, Stryker MP. Rapid extragranular plasticity in the absence of thalamocortical plasticity in the developing primary visual cortex. Science. 2000;287:2029–2032. doi: 10.1126/science.287.5460.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tudor M, Akbarian S, Chen RZ, Jaenisch R. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. doi: 10.1073/pnas.242566899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann M, Queisser G, Eder A, Wiegert JS, Bengtson CP, Hellwig A, Wittum G, Bading H. Synaptic activity induces dramatic changes in the geometry of the cell nucleus: interplay between nuclear structure, histone H3 phosphorylation, and nuclear calcium signaling. J Neurosci. 2009;29:14687–14700. doi: 10.1523/JNEUROSCI.1160-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong RO, Ghosh A. Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci. 2002;3:803–812. doi: 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, Ruan X, Ruan Y, Sun YE. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proceedings of the National Academy of Sciences. 2010;107:18161–18166. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, Thatcher KN, Farnham PJ, LaSalle JM. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proceedings of the National Academy of Sciences. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.