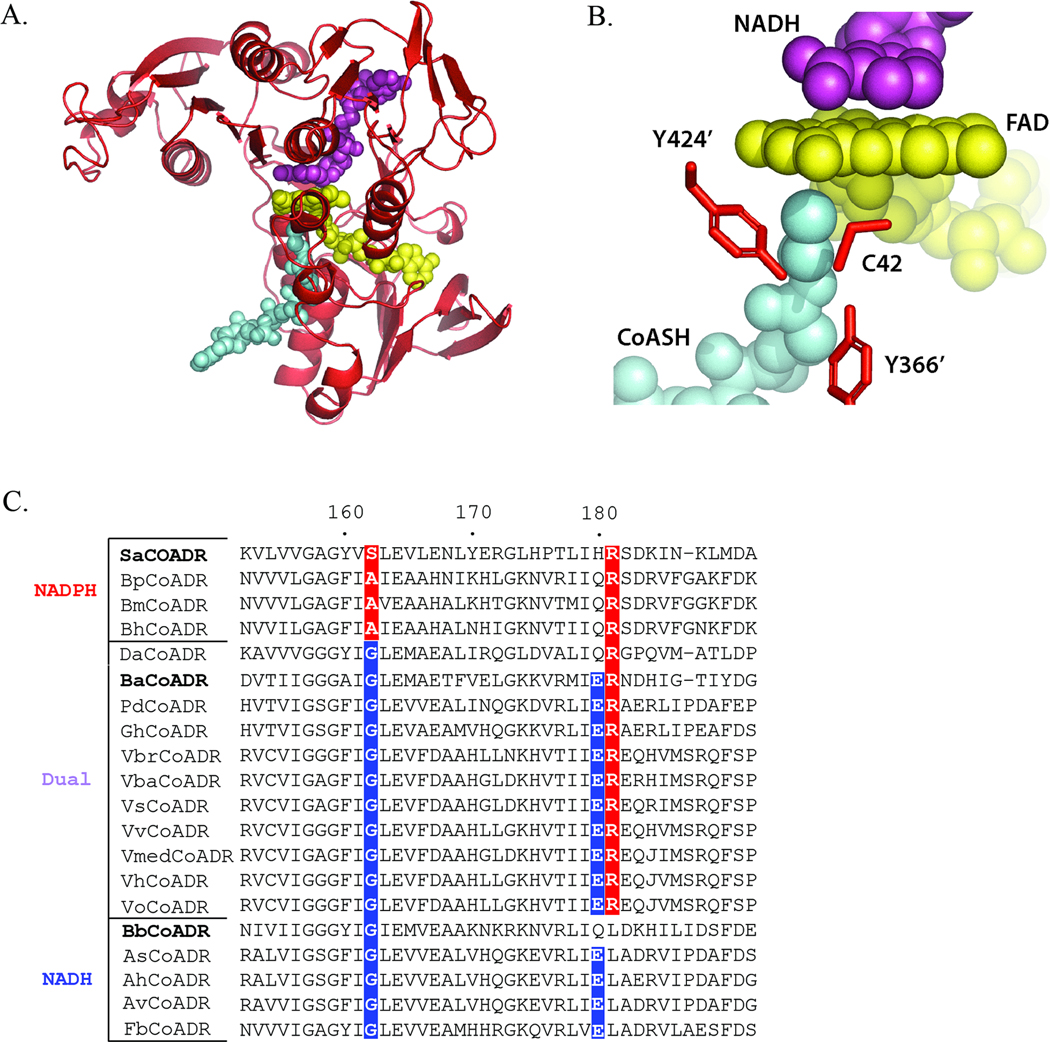
Figure 1.
(A) A monomer of a Bb CoADR homology model generated based on the structure of the reduced Ba CoADR-NADH complex (Wallen et al., 2008). CoASH, FAD, and NADH are colored cyan, yellow, and magenta, respectively. The orientation is the same as Figure 3 published in (Wallenet al., 2008). PDB code for the Ba CoADR structure used is 3CGD. (B) The predicted active site of Bb CoADR. Bb CoADR residues [Cys 42 (chain A) and Tyr366’ and Tyr424’ (chain B)] are indicated. CoASH, FAD, and NADH are colored the same as in (B).(C) Sequence alignment for the NAD(P)H-binding motifs of annotated CoADRs from Gram negative bacteria and spirochetes. Sequence numbering corresponds to Ba CoADR. Gly at position 162 and Asp or Glu at position 180 are favored for NADH-specificity, while Ala and Arg residues at positions 162 and 181, respectively, contribute to NADPH-specificity (Karplus and Schulz, 1989; Stehle et al., 1993; Wallen et al., 2008). A hybrid sequence of Glu180 and Arg181 is characteristic of dual NADH and NADPH specificity (Wallen et al., 2008). Enzymes are grouped according to predicted NAD(P)H substrate preference; positions important for NADPH- (red) or NADH- (blue) specificity are indicated. CoADRs with experimentally-determined pyridine nucleotide specificity are indicated in bold. Bp, Br. pilisicoli ; Bm, Br. murdochii ; Bh, Br. hyodysenteriae ; Da, D. acetoxidans ; Pd, Photobacterium damselae ; Gh, Grimontia hollisae ; Vbr, Vibrio brasiliensis ; Vba, V. bacterium ; Vs, V. splendidus ; Vv, V. vulnificus ; Vmed, V. sp. MED22; Vh, V. harveyi ; Vo, V. orientalis ; As, A. salmonicidia ; Ah, A. hydrophila ; Av, A. veronii ; Fb, F. balearica.