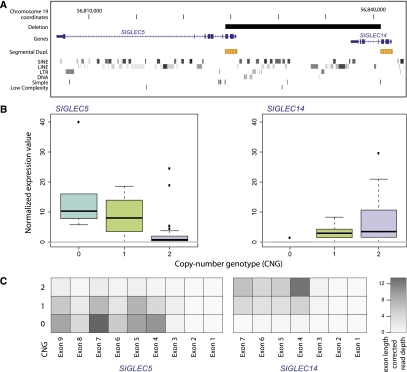
Figure 4.
Detailed expression analysis for a CNV-associated eQTL involving a polymorphic fusion gene. The expression of the adjacent genes SIGLEC5 and SIGLEC14 is affected by a deletion that partially disrupts both genes, resulting in the “SIGLEC14/5” gene hybrid. (A) SIGLEC14/5, which displays >99.8% coding sequence identity with SIGLEC5, encompasses exons 1–3 as well as upstream regulatory sequences from SIGLEC14, and further contains exons 4–9 from SIGLEC5 (Supplemental Fig. 2). We identified a CNV-associated eQTL involving the biallelic deletion and SIGLEC5 in both populations and analyzed the expression patterns of the locus making use of published nucleotide resolution breakpoint information for the deletion (Yamanaka et al. 2009). (B) The observed negative correlation between copy number and expression can plausibly be explained by the comparably higher expression of SIGLEC14 in the absence of the deletion, and further by the juxtaposition of SIGLEC14/5 with the SIGLEC14 upstream (promoter) region. Because SIGLEC14/5 displays high sequence identity with SIGLEC5, transcripts originating from the fusion gene were mostly mapped to the SIGLEC5 gene locus. Although the fusion gene is not represented in the reference genome, RNA-seq-based expression measurements at the locus were allele-specific in the presence of the homozygous reference allele (copy number [CN] = 2), and also in the presence of the homozygous deletion (CN = 0). Namely, displayed read counts for CN = 0 stem from SIGLEC14/5; for CN = 2, depicted read counts were unambiguously assigned to either SIGLEC14 or SIGLEC5 (the latter is expressed at very low level). By comparison, for CN = 1, we measured a mixture of reads originating from all three genes. (C) Read counts are shown in the form of a heat map for YRI samples, based on the raw number of RNA-seq reads that uniquely map to the reference genome at SIGLEC5 and SIGLEC14 loci. Few reads could be uniquely mapped into exons 1–3, since these fall into a segmental duplication and thus lack unique sequence. We found the deletion genotype and the expression of SIGLEC14 to be positively correlated (Spearman rank correlation P-value < 0.003 for YRI and < 0.05 for CEU), as expected as the deletion disrupts most transcript sequence unique to SIGLEC14.