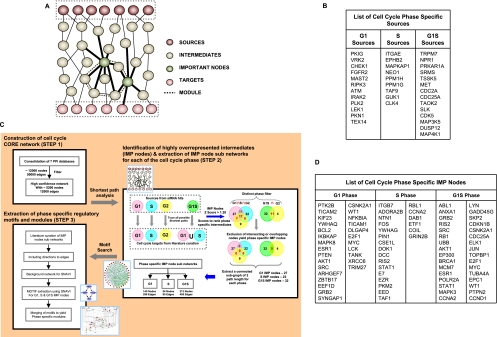
Figure 2.
A systems approach to the analysis of cell cycle regulation and the identification of phase-specific IMP nodes. (A) The rationale underlying the examination of source to target subnetworks in order to identify key intermediate effectors of cell cycle perturbation. Here the sources denote the hits identified by the siRNA, while the targets denote the key molecules involved directly in progression of cell cycle and growth. (B) The “source” characterization of the hits obtained in the siRNA screen, based on the phase-specific effects obtained. The genes described here are the human orthologs of the murine counterparts (see text). The flowchart in panel C provides a stepwise summary of the in silico methodologies used for eventual identification of the phase-specific regulatory modules. STEP 1 describes the stages involved in delineation of the core network, whereas STEP 2 illustrates the subsequent analysis of this core network to extract the phase-specific IMP node subnetworks. Finally, STEP 3 depicts the stages through which an analysis of the IMP node subnetworks eventually yielded the phase-specific modules. These individual steps are described in detail in the Supplemental Experimental Procedures. (D) The IMP nodes eventually identified through this network analysis for regulation of the G1, S, and G1S phases.