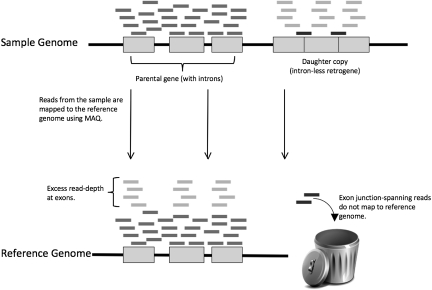
Figure 1.
Mapping reads from a genome containing a polymorphic retrocopy to a reference genome. The black line at the top represents a chromosome in a sample genome. Gray boxes represent exons within a gene, with the spaces in between representing introns. The gray boxes on the right with no introns in between them represent a retrogene derived from a parental gene (located downstream in this example). The short bars appearing above the two gene copies represent reads derived from the sample chromosome. The black line at the bottom represents the same chromosome in the reference genome to which these reads are mapped. Note that reads derived from the parental copy of the gene are mapped to the proper location, while reads from the retrocopy (light gray) are mapped to the exons of the parental copy, resulting in elevated read-depth. Also note that the reads crossing exon–exon boundaries in the retrocopy (dark gray) are not mapped to the reference genome. Our method to detect retroCNVs involves finding these reads by searching all unmapped reads against a database of exon–exon junctions.