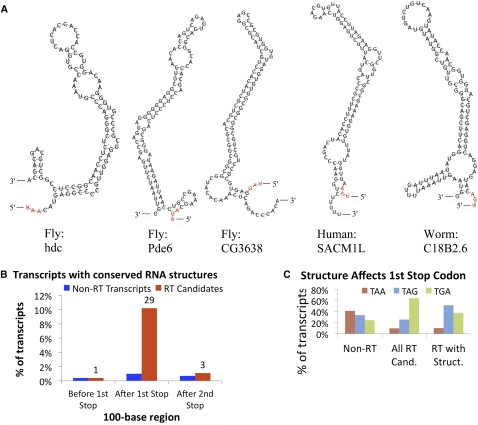
Figure 6.
RNA structures associated with readthrough genes. (A) Fly, human, and worm examples of conserved, stable RNA structures predicted in the 100-nt regions downstream from (and including) candidate readthrough stop codons. The stop codon is highlighted in red. Twenty-nine structures were found in D. melanogaster, one in human, and one in C. elegans. The stem–loop in hdc was previously found to trigger readthrough. (B) Across 283 Drosophila readthrough candidates (red bars), 10% (n = 29) showed predicted structures in the 100-nt region downstream from the first stop codon compared with only 1% for non-readthrough transcripts (blue bars). The enrichment is exclusively found downstream from the first stop codon, with only one readthrough candidate showing a predicted structure in the 100 nt upstream of the first stop codon and three in the 100-nt downstream from the second stop codon, suggesting potential interactions with the ribosome during reading of the readthrough stop position. (C) Readthrough stop codon usage among readthrough candidates with and without predicted structures and non-readthrough genes. Although most readthrough candidates use TGA, readthrough candidates with structures show a preference for TAG, suggesting that a leaky stop codon context might not be necessary for readthrough in the presence of RNA structures.