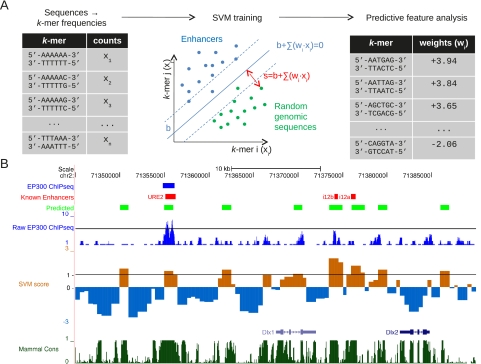
Figure 1.
Overview of our methodology. (A) k-mer frequencies are calculated for each of the EP300-bound and negative genomic training sequences. These feature vectors (x1,…,xn) are used to find SVM weights, w, which most accurately separate the positive (enhancer) and negative (genomic) training sets. (B) These weights are used to predict genome-wide enhancers (light green), based on their SVM score. (Brown) positive, (blue) negative. A well-studied region around Dlx1 and Dlx2 is shown here, both known to be expressed in the forebrain. While the predicted enhancers often overlap the training EP300 set (blue), novel enhancers are also predicted and often identify previously experimentally verified enhancers (red) absent from the EP300 training set. The predicted enhancers also preferentially occur in conserved nonexonic regions (dark green) and regions enriched in EP300 signal (dark blue).