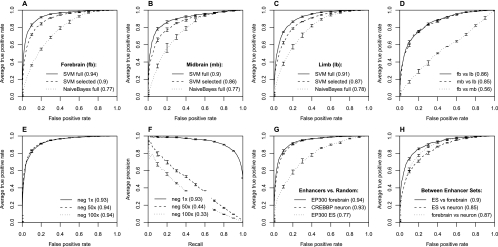
Figure 2.
Classification results on each tissue-specific enhancer set. (A) Classification of forebrain enhancers vs. random genomic sequences. (B) Classification of midbrain enhancers vs. random genomic sequences. (C) Classification of limb enhancers vs. random genomic sequences. Each graph in A, B, and C compares an SVM trained on the full set of 6-mers (solid), the top 100 selected 6-mers (dashed), and an alternative Naive Bayes classifier (dotted). Each curve is an average of five cross-fold validations on a reserved test set; error bars denote one standard deviation over the five cross-fold validation sets. Numbers in parentheses indicate the area under each ROC curve (auROC) for overall comparison. Both the full SVM and SVM with selected features perform very well and significantly better than Naive Bayes. Individually, each tissue-specific set can be accurately discriminated from nonenhancer genomic sequences. (D) Classification of specific tissues vs. other tissues. Forebrain (fb) and midbrain (mb) can be accurately discriminated from limb (lb) but not from each other (fb vs. mb), indicating common or overlapping modes of regulation. (E) Classification ROC curves for forebrain enhancers vs. random genomic sequences for larger negative set sizes. (F) Precision-recall curves for forebrain enhancers vs. random sequences corresponding to the ROC curves and negative sets in E; numbers in parentheses are auPRC. (G) Classification of EP300 forebrain enhancers, neuronal stimulus-dependent enhancers (CREBBP neuron), and mouse embryonic stem cell enhancers (EP300 ES) vs. random genomic sequence. Although the embryonic stem cell data set is somewhat less accurately classified, our SVMs successfully discriminate EP300 or CREBBP bound regions from random sequences. (H) Classification of EP300 fb, CREBBP neuron, and EP300 ES data sets vs. each other is also robust.