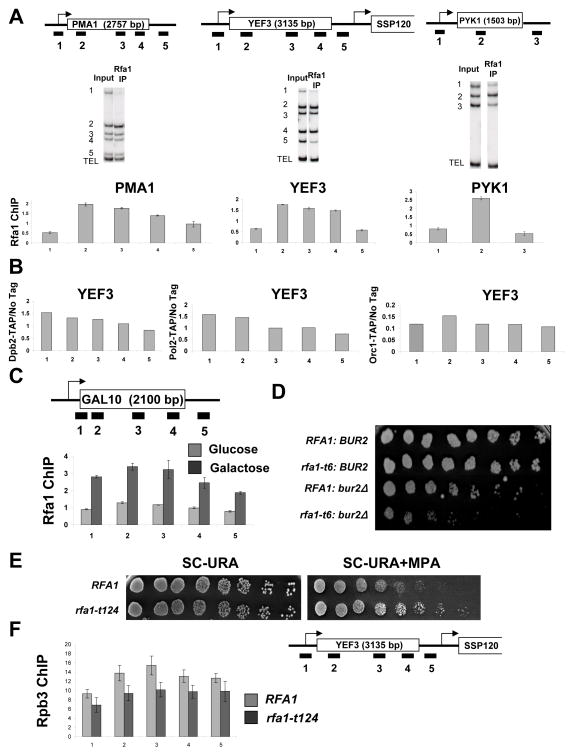
Figure 5. RPA associates with active genes.
(A) ChIP was performed using an antibody against the Rfa1 subunit of RPA. Cop-precipitated DNA was analyzed across the PMA1, YEF3, and PYK1 genes as described in Figure 2. The values shown represent the averages and standard errors (bars) from three independent experiments. (B) ChIP was performed using strains expressing TAP-tagged Dpb2 or Pol2, (components of DNA Polymerase (II) ε), or Orc1 (component of the replication origin recognition complex). DNA coprecipitated with the TAP-tagged proteins was analyzed by PCR for regions across the YEF3 gene. (C) Cells were grown in YP-Glucose or YP-Galactose to an OD ~0.5, and Rfa1 ChIP was performed as above except testing for regions across the GAL10 gene. Quantitation of triplicate experiments is as in part A. (D) Strains with the indicated genotypes were spotted in 3-fold dilutions on Synthetic Complete (SC) + 5-Fluoroorotic Acid to remove an RFA1-URA3 covering plasmid. Plates were incubated for 6 days at 30°C. (E) The indicated isogenic strains were spotted in 3-fold dilutions on SC plates lacking uracil (3 days growth shown) or SC-URA plates containing MPA (4 days). (F) ChIP of the Rpb3 subunit of RNApII at the YEF3 gene was performed in strains with wild-type Rfa1 (RFA1) or expressing the rfa1-t124 allele grown in galactose. Quantitation of triplicate experiments is as in part A.