Figure 2.
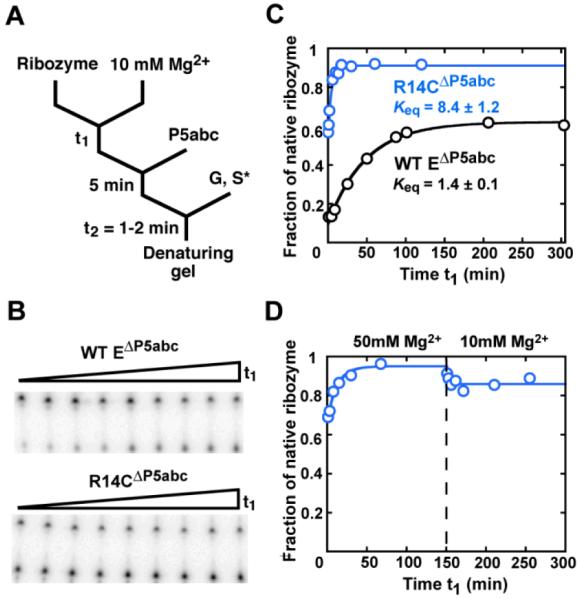
Increased specificity for native folding in the R14CΔP5abc ribozyme. (A) RNA was folded in Mg2+ for time t1, and then P5abc was added, followed by S* and guanosine (G). Reactions were incubated long enough to give complete S* cleavage by the native ribozyme (time t2, 1 min for the wild-type ribozyme and 1.75 min for mutants with modestly reduced cleavage rates; see Methods). (B) Gel showing approach to equilibrium of native and misfolded conformations for the wild-type EΔP5abc (top) and R14CΔP5abc (bottom) ribozymes. Time t1 was varied from 1 min to >250 min. (C) Folding progress curves for R14CΔP5abc (blue) and EΔP5abc (black) in side-by-side reactions. Rate constants for the approach to equilibrium were 0.25 ± 0.01 min−1 and 0.018 ± 0.001 min−1 for R14CΔP5abc and EΔP5abc, respectively (average ± standard error). The latter result is the same within error as reported previously (21) Fractions of native ribozyme in this and other figure s are calculated by dividing the fraction. of S* cleaved during t2 by the corresponding fraction for fully-native ribozyme formed by extended incubation with P5abc (>0.8, see Methods). (D) Approach to equilibrium of R14CΔP5abc upon folding at 50 mM Mg2+ and subsequent dilution to standard conditions (10 mM Mg2+).
