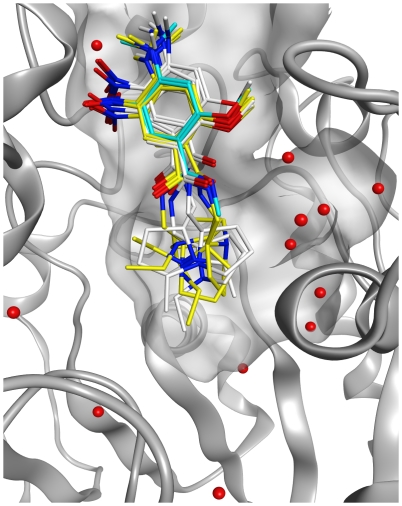
Figure 7. Predicted binding poses of both C5685 enantiomers by molecular docking simulations.
Poses of C5685 (R) and C5685 (S) that were generated using the modified parameter settings in Glide are shown with carbons colored white and yellow, respectively. The part of C5685 as modeled in the X-ray crystal structure is shown with carbons colored in cyan, and the 12 conserved water molecules that were included in the docking simulations are indicated in red.