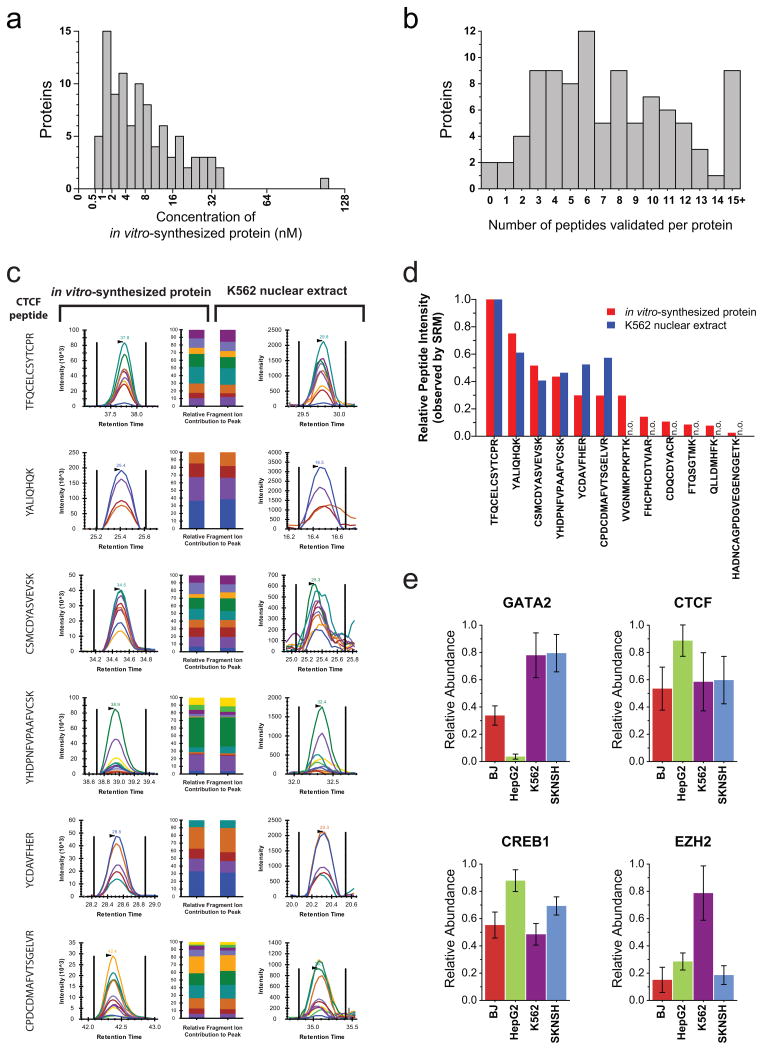
Figure 2. Targeted assays can be efficiently developed using in vitro synthesized proteins and applied to measure proteins in vivo.
(a)The absolute quantity of each in vitro-synthesized protein sample, as measured using a tryptic peptide contained within the c-terminal schistosomal GST tag. (b) The number of peptides per protein empirically assessed with salient features to accurately detect and quantify the target proteins (peptides with a quality score of either 1 or 2). (c) Proteotypic peptides identified using in vitro-synthesized CTCF were monitored in K562 nuclear extracts. The relative contribution of each fragment ion to each peptide peak is displayed as different colors. (d)For each proteotypic peptide from CTCF, the relative signal intensity observed using in vitro synthesized protein is displayed alongside the relative signal intensity observed using K562 nuclear extract Peptides not observed (n.o.) in K562 nuclear extracts are indicated. (e) The measured relative abundance of four transcription factors between the fibroblast (BJ), hepatic carcinoma (HepG2), erythroleukemia (K562) and neuroblastoma (SKNSH) human cell lines. Data points are mean ± s. d. (n = 6).