Abstract
Background
Helicobacter pylori is a persistent colonizer of the human gastric mucosa, which can lead to the development peptic ulcer disease and gastric adenocarcinomas. However, H. pylori can asymptomatically colonize a host for years. One factor that has been hypothesized to contribute to such persistence is the production of Lewis (Le) antigens in the lipopolysaccharide layer of the bacterial outer membrane as a form of molecular mimicry, since humans also express these antigens on their gastric mucosa. Humans and H. pylori both are polymorphic for Le expression, which is driven in H. pylori by variation at the Le synthesis loci. In this report we sought to characterize Le genotypic and phenotypic variation in geographically diverse H. pylori isolates.
Materials and Methods
From patients undergoing endoscopy in 29 countries, we determined Le phenotypes of 78 H. pylori strains, and performed genotyping of the galT and β-(1,3)galT loci in 113 H. pylori strains.
Results
Le antigen phenotyping revealed a significant (p <0.0001) association between type 1 (Lea and Leb) expression and strains of East-Asian origin. Genotyping revealed a significant correlation between strain origin and the size of the promoter region upstream of the Le synthesis gene, galT (p <0.0001).
Conclusion
These results indicate that the heterogeneity of human Le phenotypes are reflected in their H. pylori colonizing strains, and suggest new loci that can be studied to assess variation of Le expression.
Introduction
Helicobacter pylori are Gram negative, microaerophilic bacteria that colonize the human stomach. This persistent colonization has been linked to gastric ulcers, gastric adenocarcinoma (1), and mucosa-associated lymphoid tissue (MALT) lymphoma (2). As a result of these serious consequences, currently most treatment protocols for peptic ulcer disease include eradication of H. pylori as part of their regimens (3). However, colonization with H. pylori can go undetected for decades, and may have some early-in-life benefits (4, 5); how H. pylori is able to persist within the host for such long periods is not clearly understood.
Lewis (Le) antigens are cell-surface fucosylated oligosaccharides that are expressed in both humans (6) and H. pylori (7–10). It has been hypothesized that H. pylori presents these antigens within its lipopolysaccharide (LPS) layer as a form of molecular mimicry, perhaps aiding in niche adaptation and evasion of host immune responses (11–17). Type 2 antigens (Lex and Ley) are most commonly expressed (~85 % of strains, (18, 19)), while type 1 antigens (Lea and Leb) are expressed in less than 5% of collections of H. pylori strains studied (18, 19). Both observational (16) and experimental (15, 20) studies have demonstrated a relationship between host and bacterial Le phenotype, suggesting that the host Le phenotype selects for bacterial Le phenotype. Furthermore, Le expression in H. pylori appears to correlate with the geographic origin of its human host; North American and European strains predominantly express type 2 Le antigens only, while type 1 Le antigen expression, along with simultaneous type 2 Le antigen expression, appears to be more prevalent in Asian and the limited numbers of South American strains studied (7–9, 18, 21–23).
Lewis antigens are synthesized from a common precursor, N–acetylglucosamine, which is galactosylated in the type 1 or 2 synthesis pathways by β-(1,3)galT or GalT, respectively (15, 24–26). These precursor disaccharides then can be mono-fucosylated to form the trisaccharides Lex and Lea (27–31), or difucosylated to form the tetrasaccharides Ley or Leb (28, 32).
Recently it has been reported that the β-(1,3)galT upstream homolog, jhp0562, is a potential marker for peptic ulcer disease (PUD) in children (33, 34), and its presence has been associated with the presence of H. pylori proteins (e.g. CagA) associated with high intensity host interactions (34). While only present in some strains of H. pylori, in those strains that possess a copy of jhp0562, mutagenesis has shown that this gene is essential for synthesis of all Le antigens (35). However, strains that naturally lack jhp0562 also have the ability to produce type 2 antigens (34, 36, 37). In this study, we aimed to further elucidate the variation at the β-(1,3)galT and galT loci amongst H. pylori strains isolated from different human populations, especially in relation to Lewis antigen expression, and to examine the relationship between H. pylori Le antigen polymorphisms and the geographic origin of the host.
Methods
Patient population
The H. pylori-positive population consisted of patients undergoing upper gastrointestinal (UGI) endoscopy as part of routine treatment at the New York Harbor (Manhattan) VA Medical Center, Bellevue Hospital Center, and New York Downtown Hospital as well as patient samples isolated in other parts of the world, including Latin American countries and Europe, collected between 1984 and 2003 (Table S1). H. pylori-positive patients were defined on the basis of positive H. pylori cultures obtained from gastric biopsies. Patients were separated into two groups, “ East Asian” and “Non-East Asian”, with the former defined as having an East-Asian ethnicity, and which for the purposes of this study included patients originating in China, Taiwan, Japan, Hong Kong, Burma, Malaysia, and Thailand. The “Non-East Asian” group included all other patients (n = 116 subjects).
Bacterial strains and growth conditions
H. pylori strains were isolated from biopsy samples by plating on Skirrow’s medium (BBL Microbiology Systems, Cockeysville, MD), and grown at 37°C under microaerobic conditions. Frozen stocks were maintained at −80°C in Brucella broth (BB) with 15% glycerol. Strains were routinely grown on 5% sheep’s blood agar (BBL Microbiology Systems, Cockeysville, MD) at 37°C and 5% CO2, or in culture jars under microaerobic conditions.
Determination of Lewis antigen phenotypes
H. pylori Le antigen phenotypes were determined by ELISA using monoclonal antibodies to Lea, Leb, Lex, or Ley (Signet Laboratories, Inc., Dedham, MA) by methods described (19). Optical densities (OD) at 410 nm were determined on a microplate reader (MRX; Dynatech Laboratories, Inc., Chantilly, VA). Two previously defined strains, JP26, a wild-type Leb-positive strain isolated in Japan, and 99-8, a Lea-positive strain from our collection were included as controls (15). Corrected OD values were determined by obtaining the mean of the OD values from three wells per sample and subtracting the blank (Escherichia coli strain HB101). Lewis antigen expression was considered to be positive if the OD values were greater than 0.10. A subset (n=78) of the strains analyzed in this study were Le antigen phenotyped, based on our ability to recover a sufficient number of viable cells from frozen stock for ELISA.
PCR analysis of the β-(1,3)galT locus and galT promoter region
H. pylori strains were harvested from a single plate in 1.0 ml sterile phosphate buffered saline (PBS, pH 7.4), cells pelleted, and genomic DNA was extracted with the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA). Purified DNA was used as template to screen for the presence of jhp0562 using primers Jhp0561(+419)F and Jhp0564( −10)R (15), flanking the jhp0562-β-(1,3)galT locus, and primers Jhp0562(+211)F (15) and jhp0562 (+577)R CATGCGTTGAGTAATAGCTTTTTTG, specific for jhp0562. To determine the relative size of the galT upstream region, primers 2(1,4)galT(−391)F and Gal(1,4)R(+50) were used (15).
Statistical analysis
Either Chi-squared analysis or Fisher’s exact test was used as appropriate based on cell size, with p < 0.05 considered significant.
Results
Prevalence of jhp0562 in H. pylori isolates of East Asian and Non-East Asian origin
To determine the prevalence of the presence of jhp0562 at the β-(1,3)galT locus, PCR analysis of 111 H. pylori isolates was performed using primers flanking the region as well as with primers specific for jhp0562 (Figure 1). Isolates harboring both genes yield a band of ~2.6kb, while isolates lacking jhp0562 yield a band of ~1.5kb (Figure 1A). In some isolates producing the larger 2.6kb band, a faint band 1.5kb band is also observed (Figure 1A), which can be attributed to intragenomic recombination between the two homologous alleles (35). The results in Figure 1A were confirmed by a jhp0562-specific PCR (Figure 1B). The jhp0562 allele was detected in 56/68 (82.4%) of Non-Asian strains, while 41/43 (95.3%) of East-Asian strains were jhp0562 positive (Table 1). These results show that the majority of H. pylori strains tested harbor jhp0562, but jhp0562 status trended toward significance between the two groups (p = 0.075, Table 1).
Figure 1. PCR amplification of the jhp0562-β-(1,3)galT(jhp0563) locus.
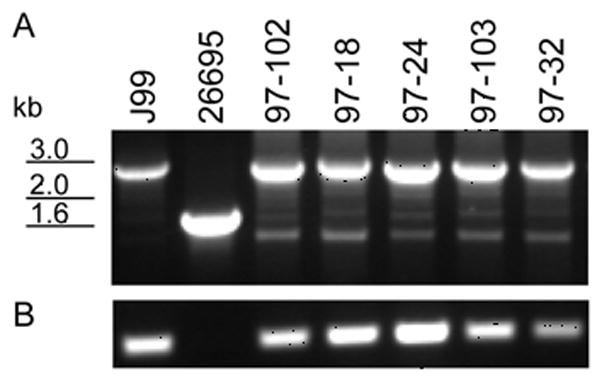
Panel A. Representative results of PCR screens for the presence of jhp0562 with primers flanking jhp0562- β-(1,3)galT. Strains harboring a copy of both jhp0562 and β-(1,3)galT produce a band of ~2.6 kb, while strains lacking jhp0562 amplify a band of ~1.5 kb. Whole genomic sequencing of strains J99 and 26695 indicate that strain J99 possesses both genes, whereas 26695 only possess β-(1,3)galT (37, 39). Panel B. jhp0562-specific PCR. Only H. pylori strains that contain a copy of jhp0562 yield a band in this assay.
Table 1.
Le antigen phenotypes and genotypes of H. pylori strains, according to their geographic origin.
| Phenotypes (n=78) | East Asian | Non-East Asian | Total | p-valuea |
|---|---|---|---|---|
| Le negative | 3 | 8 | 11 | |
|
| ||||
| Le positive | ||||
| Type 1 only | 0 | 0 | 0 | - |
| Type 2 only | 23 | 31 | 54 | - |
| Type 1 & 2 | 13 | 0 | 13 | <0.0001 |
|
| ||||
| Genotypes
| ||||
| jhp0562 + (n =111) | 41 | 56 | 97 | |
| jhp0562 − | 2 | 12 | 14 | 0.075 |
|
| ||||
| galT promoter (n=113) | ||||
| Small | 33 | 3 | 36 | - |
| Medium | 7 | 7 | 14 | 0.0026b |
| Large | 2 | 61 | 63 | <0.0001c |
Fisher’s exact test
In relation to the number of small galT promoter regions.
In relation to the number of strains that have smaller galT promoter sizes (small and medium), compared to the number of strains that have large galT promoter regions.
Prevalence of type 1 Le antigen between East Asian and Non-East Asian-derived H. pylori populations
Le antigen phenotyping by ELISA was performed on 78 strains: 39 non-Asian and 39 East Asian strains. No strains of non-East Asian origin were positive for type 1 Le antigens, while 13 East Asian strains expressed type 1 Le antigens, a difference that was significant (p <0.0001, Table 1).
Correlation of type 1 Le antigen expression with jhp0562 status
All 11 H. pylori strains that expressed type 1 Le antigens that were screened for the presence of jhp0562 carried the gene. However, there was no correlation between the presence of jhp0562 and type 1 expression (p=0.58).
Association between ethinic origin of H. pylori strains and galT promoter region
Although the experimentally defined transcripton start site for galT begins 31–33 nucleotides before the galT translational start site [(38), Figure 2], the promoter region of galT varies in size amongst H. pylori strains [e.g. 26695 and J99 (37, 39), Figure 2]. Using a forward primer 391nucleotides upstream of the galT start codon and a reverse primer annealing 50 nucleotides downstream of the start codon produces alleles of three different sizes, termed “small”, “medium” and “large” among isolates characterized in this study (Figure 3). A PCR screen with these primers was performed on 115 H. pylori strains. All strains produced a single band, with the exception of two strains, 02-363 and 03-151, which produced two bands, one small and one large, suggesting that these strains represent mixed populations (Figure 3). Chi-squared analysis revealed that strains of Non-East Asian origin most commonly possess a “large” galT promoter region, while Asian strains were significantly (p < 0.0001) associated with “small” galT promoter regions (Table 1).
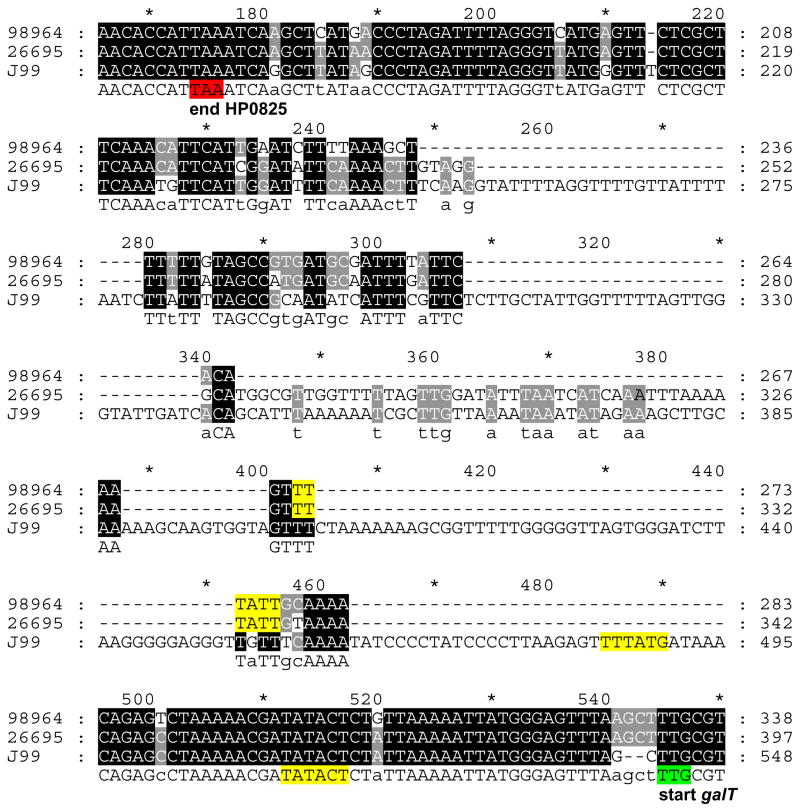
Figure 2. Nucleotide alignment of the region upstream of galT in three representative H. pylori strains.
The strains are 98-964, 26695 and J99, with small, medium, and large galT upstream regions, respectively, as detected by PCR (Figure 3). The stop codon of the gene upstream of galT, HP0825 is highlighted in red, while the start codon of galT is shown in green. Predicted −35 and −10 sequences (38, 61), are highlighted in yellow.
Figure 3. PCR screen of the galT promoter region in H. pylori strains.
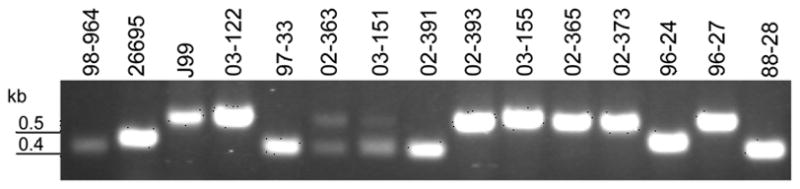
Results of representive PCR amplification of the galT promoter region. Three different promoter sizes were detected: small, as in control strain 98-964, medium, as in strain 26695; and large, as in control strain J99.
Discussion
H. pylori strains are varied in their expression of Le antigens (7–10, 13, 15, 16, 19–22, 24, 28, 40–45), and multiple genetic mechanisms to create such variation have been described (15, 24, 32, 40, 41, 44, 46–50). The results of the genotypic and phenotypic screens in this study demonstrate the high levels of variation observed at the sites that are critical to Le antigen synthesis and identify new loci that are relevant to variation. Our results showed that H. pylori strains of East-Asian origin are more likely to express type 1 Le antigens on their LPS than strains of North American or European origin, confirming and extending prior studies (21, 22). In addition, it has been shown that H. pylori strains of South American origin express type 1 Le antigens more often than Western strains (23), suggesting that there are global distribution patterns of H. pylori Le antigen phenotypes. These patterns could reflect the prevalence of particular Le expression profiles on the gastric epithelia of different human populations (51), which would further support the hypothesis that H. pylori strains in which Le expression matches that of their host are selected (15, 16, 20). However, to our knowledge no studies comparing human and bacterial Le expression in geographically diverse isolates have been performed.
Our results showed no correlation between type 1 Le expression and the presence of jhp0562. However, previous work in our lab has shown that in strains in which it is present, jhp0562 is essential for production of all Le antigens (35). However, from prior studies (34), and our current work, a substantial (37.6% and 9.7%, respectively) proportion of clinical isolates lack jhp0562 but express type 2 Le antigens. Thus, there appears to be a fundamental difference between strains carrying jhp0562 in which it is essential for Le antigen synthesis, and those without it, which can at least produce type 2 antigens. The mechanisms that underlie this difference are not known, although it is possible that another Le antigen synthesis gene is compensating for the lack of jhp0562 in strains that do not have a copy of this gene, thus allowing for type 2 Le antigen production in these strains. The presence of jhp0562 is correlated with PUD in children (33, 34), as well as other H. pylori virulence genes, including vacA s1, babA, homB, oipA, hopQ I, and those on the cag PAI, (34). Thus, although jhp0562 is not an essential gene, its association with PUD and host-associated genes suggests a role in host interaction, perhaps by aiding in colonization and niche adaptation via its role in Le antigen synthesis.
PCR analysis of the galT promoter region revealed major alleles of three different sizes, “small”, “medium” and “large”, with East-Asian strains strongly associated with the small allele and Non-East Asian (Western) strains predominantly harboring the large allele. However, the predicted transcriptional start site (38) is conserved amongst the alleles (Figure 2). Thus, further studies are necessary to determine whether size variation of the promoter region affects transcriptional activity of galT. Additionally, two strains showed multiple bands, indicating a mixed population of cells within the isolate, suggesting that the patient was colonized with multiple H. pylori strains or a single strain with clonal variants, phenomena that have been observed previously (43, 52–55). Alternatively, these multiple bands could be the result of intragenomic or intergenomic recombination, commonly observed in H. pylori at various loci (35, 56–58), including between Le synthesis genes (44). Harboring alleles of different sizes within a particular population could be advantageous by allowing for variable expression of Le antigens, thus aiding in adaptation to different microniches within the host (43, 55, 56, 59).
Our results have shown that distinct H. pylori genotypes and phenotypes related to Le antigens are associated with geographic origins of the strain. Such extensive phenotypic variation perhaps reflects H. pylori’s ability to mimic the Le phenotype of its host as a means of niche adaptation and survival (15, 47). Divergence of other Le antigen synthesis genes between European and East-Asian strains further supports this hypothesis (36, 60). These results provide the framework for future studies to investigate the relationship between host and H. pylori Le antigen phenotypes, and how such phenotypic variation may contribute to H. pylori’s ability to persistently colonize the human gastric mucosa.
Supplementary Material
Acknowledgments
This work was supported in part by RO1GM63270 and 2T32 AI007180 from the National Institutes of Health, by the Medical Research Service of the Department of Veterans Affairs, and by the Diane Belfer Program for Human Microbial Ecology. The authors have no conflicting financial interests related to this work.
References
- 1.Peek RM, Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002 Jan;2(1):28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 2.Stolte M, Bayerdorffer E, Morgner A, Alpen B, Wundisch T, Thiede C, et al. Helicobacter and gastric MALT lymphoma. Gut. 2002 May;50(Suppl 3):III19–24. doi: 10.1136/gut.50.suppl_3.iii19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costa F, D'Elios MM. Management of Helicobacter pylori infection. Expert Rev Anti Infect Ther. 2010 Aug;8(8):887–92. doi: 10.1586/eri.10.75. [DOI] [PubMed] [Google Scholar]
- 4.Atherton J, Blaser M. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. The Journal of clinical investigation. 2009;119(9):2475–87. doi: 10.1172/JCI38605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y, Blaser M. Helicobacter pylori colonization is inversely associated with childhood asthma. The Journal of infectious diseases. 2008;198(4):553–60. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakamoto S, Watanabe T, Tokumaru T, Takagi H, Nakazato H, Lloyd KO. Expression of Lewisa, Lewisb, Lewisx, Lewisy, siayl-Lewisa, and sialyl-Lewisx blood group antigens in human gastric carcinoma and in normal gastric tissue. Cancer Res. 1989 Feb 1;49(3):745–52. [PubMed] [Google Scholar]
- 7.Aspinall GO, Monteiro MA. Lipopolysaccharides of Helicobacter pylori strains P466 and MO19: structures of the O antigen and core oligosaccharide regions. Biochemistry. 1996 Feb 20;35(7):2498–504. doi: 10.1021/bi951853k. [DOI] [PubMed] [Google Scholar]
- 8.Aspinall GO, Monteiro MA, Pang H, Walsh EJ, Moran AP. Lipopolysaccharide of the Helicobacter pylori type strain NCTC 11637 (ATCC 43504): structure of the O antigen chain and core oligosaccharide regions. Biochemistry. 1996 Feb 20;35(7):2489–97. doi: 10.1021/bi951852s. [DOI] [PubMed] [Google Scholar]
- 9.Monteiro MA, Appelmelk BJ, Rasko DA, Moran AP, Hynes SO, MacLean LL, et al. Lipopolysaccharide structures of Helicobacter pylori genomic strains 26695 and J99, mouse model H. pylori Sydney strain, H. pylori P466 carrying sialyl Lewis X, and H. pylori UA915 expressing Lewis B classification of H. pylori lipopolysaccharides into glycotype families. Eur J Biochem. 2000 Jan;267(2):305–20. doi: 10.1046/j.1432-1327.2000.01007.x. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro MA, Chan KH, Rasko DA, Taylor DE, Zheng PY, Appelmelk BJ, et al. Simultaneous expression of type 1 and type 2 Lewis blood group antigens by Helicobacter pylori lipopolysaccharides. Molecular mimicry between h. pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. J Biol Chem. 1998 May 8;273(19):11533–43. doi: 10.1074/jbc.273.19.11533. [DOI] [PubMed] [Google Scholar]
- 11.Negrini R, Savio A, Poiesi C, Appelmelk BJ, Buffoli F, Paterlini A, et al. Antigenic mimicry between Helicobacter pylori and gastric mucosa in the pathogenesis of body atrophic gastritis. Gastroenterology. 1996 Sep;111(3):655–65. doi: 10.1053/gast.1996.v111.pm8780570. [DOI] [PubMed] [Google Scholar]
- 12.Sherburne R, Taylor DE. Helicobacter pylori expresses a complex surface carbohydrate, Lewis X. Infect Immun. 1995 Dec;63(12):4564–8. doi: 10.1128/iai.63.12.4564-4568.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran AP, Knirel YA, Senchenkova SN, Widmalm G, Hynes SO, Jansson PE. Phenotypic variation in molecular mimicry between Helicobacter pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. Acid-induced phase variation in Lewis(x) and Lewis(y) expression by H. Pylori lipopolysaccharides. J Biol Chem. 2002 Feb 22;277(8):5785–95. doi: 10.1074/jbc.M108574200. [DOI] [PubMed] [Google Scholar]
- 14.Appelmelk BJ, Simoons-Smit I, Negrini R, Moran AP, Aspinall GO, Forte JG, et al. Potential role of molecular mimicry between Helicobacter pylori lipopolysaccharide and host Lewis blood group antigens in autoimmunity. Infect Immun. 1996 Jun;64(6):2031–40. doi: 10.1128/iai.64.6.2031-2040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pohl MA, Romero-Gallo J, Guruge JL, Tse DB, Gordon JI, Blaser MJ. Host-dependent Lewis (Le) antigen expression in Helicobacter pylori cells recovered from Leb-transgenic mice. The Journal of experimental medicine. 2009;206(13):3061–72. doi: 10.1084/jem.20090683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirth HP, Yang M, Peek RM, Jr, Tham KT, Blaser MJ. Helicobacter pylori Lewis expression is related to the host Lewis phenotype. Gastroenterology. 1997 Oct;113(4):1091–8. doi: 10.1053/gast.1997.v113.pm9322503. [DOI] [PubMed] [Google Scholar]
- 17.Appelmelk BJ, Negrini R, Moran AP, Kuipers EJ. Molecular mimicry between Helicobacter pylori and the host. Trends Microbiol. 1997 Feb;5(2):70–3. doi: 10.1016/S0966-842X(96)10084-6. [DOI] [PubMed] [Google Scholar]
- 18.Simoons-Smit IM, Appelmelk BJ, Verboom T, Negrini R, Penner JL, Aspinall GO, et al. Typing of Helicobacter pylori with monoclonal antibodies against Lewis antigens in lipopolysaccharide. J Clin Microbiol. 1996 Sep;34(9):2196–200. doi: 10.1128/jcm.34.9.2196-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wirth HP, Yang M, Karita M, Blaser MJ. Expression of the human cell surface glycoconjugates Lewis x and Lewis y by Helicobacter pylori isolates is related to cagA status. Infect Immun. 1996 Nov;64(11):4598–605. doi: 10.1128/iai.64.11.4598-4605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth HP, Yang M, Sanabria-Valentin E, Berg DE, Dubois A, Blaser MJ. Host Lewis phenotype-dependent Helicobacter pylori Lewis antigen expression in rhesus monkeys. Faseb J. 2006 Jul;20(9):1534–6. doi: 10.1096/fj.05-5529fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteiro MA, Zheng P, Ho B, Yokota S, Amano K, Pan Z, et al. Expression of histo-blood group antigens by lipopolysaccharides of Helicobacter pylori strains from asian hosts: the propensity to express type 1 blood-group antigens. Glycobiology. 2000 Jul;10(7):701–13. doi: 10.1093/glycob/10.7.701. [DOI] [PubMed] [Google Scholar]
- 22.Zheng PY, Hua J, Yeoh KG, Ho B. Association of peptic ulcer with increased expression of Lewis antigens but not cagA, iceA, and vacA in Helicobacter pylori isolates in an Asian population. Gut. 2000 Jul;47(1):18–22. doi: 10.1136/gut.47.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altman E, Fernandez H, Chandan V, Harrison BA, Schuster MW, Rademacher LO, et al. Analysis of Helicobacter pylori isolates from Chile: occurrence of selective type 1 Lewis b antigen expression in lipopolysaccharide. J Med Microbiol. 2008 May;57(Pt 5):585–91. doi: 10.1099/jmm.0.47783-0. [DOI] [PubMed] [Google Scholar]
- 24.Appelmelk BJ, Martino MC, Veenhof E, Monteiro MA, Maaskant JJ, Negrini R, et al. Phase variation in H type I and Lewis a epitopes of Helicobacter pylori lipopolysaccharide. Infect Immun. 2000 Oct;68(10):5928–32. doi: 10.1128/iai.68.10.5928-5932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endo T, Koizumi S, Tabata K, Ozaki A. Cloning and expression of beta1,4-galactosyltransferase gene from Helicobacter pylori. Glycobiology. 2000 Aug;10(8):809–13. doi: 10.1093/glycob/10.8.809. [DOI] [PubMed] [Google Scholar]
- 26.Logan SM, Conlan JW, Monteiro MA, Wakarchuk WW, Altman E. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol Microbiol. 2000 Mar;35(5):1156–67. doi: 10.1046/j.1365-2958.2000.01784.x. [DOI] [PubMed] [Google Scholar]
- 27.Rasko DA, Wang G, Palcic MM, Taylor DE. Cloning and characterization of the alpha(1,3/4) fucosyltransferase of Helicobacter pylori. J Biol Chem. 2000 Feb 18;275(7):4988–94. doi: 10.1074/jbc.275.7.4988. [DOI] [PubMed] [Google Scholar]
- 28.Rasko DA, Wang G, Monteiro MA, Palcic MM, Taylor DE. Synthesis of mono- and di-fucosylated type I Lewis blood group antigens by Helicobacter pylori. Eur J Biochem. 2000 Oct;267(19):6059–66. doi: 10.1046/j.1432-1327.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- 29.Chan NW, Stangier K, Sherburne R, Taylor DE, Zhang Y, Dovichi NJ, et al. The biosynthesis of Lewis X in Helicobacter pylori. Glycobiology. 1995 Oct;5(7):683–8. doi: 10.1093/glycob/5.7.683. [DOI] [PubMed] [Google Scholar]
- 30.Ge Z, Chan NW, Palcic MM, Taylor DE. Cloning and heterologous expression of an alpha1,3-fucosyltransferase gene from the gastric pathogen Helicobacter pylori. J Biol Chem. 1997 Aug 22;272(34):21357–63. doi: 10.1074/jbc.272.34.21357. [DOI] [PubMed] [Google Scholar]
- 31.Martin SL, Edbrooke MR, Hodgman TC, van den Eijnden DH, Bird MI. Lewis X biosynthesis in Helicobacter pylori. Molecular cloning of an alpha(1,3)-fucosyltransferase gene. J Biol Chem. 1997 Aug 22;272(34):21349–56. doi: 10.1074/jbc.272.34.21349. [DOI] [PubMed] [Google Scholar]
- 32.Wang G, Rasko DA, Sherburne R, Taylor DE. Molecular genetic basis for the variable expression of Lewis Y antigen in Helicobacter pylori: analysis of the alpha (1,2) fucosyltransferase gene. Mol Microbiol. 1999 Feb;31(4):1265–74. doi: 10.1046/j.1365-2958.1999.01268.x. [DOI] [PubMed] [Google Scholar]
- 33.Oleastro M, Monteiro L, Lehours P, Megraud F, Menard A. Identification of markers for Helicobacter pylori strains isolated from children with peptic ulcer disease by suppressive subtractive hybridization. Infect Immun. 2006 Jul;74(7):4064–74. doi: 10.1128/IAI.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oleastro M, Santos A, Cordeiro R, Nunes B, Megraud F, Menard A. Clinical relevance and diversity of two homologous genes encoding glycosyltransferases in Helicobacter pylori. J Clin Microbiol. 2010 Aug;48(8):2885–91. doi: 10.1128/JCM.00401-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pohl MA, Blaser MJ. Helicobacter pylori Lewis antigen synthesis; 15th Int Workshop Campylobacter Helicobacter Related Organisms International Workshop on Campylobacter, Helicobacter, and Related Organisms, Niigata, Japan (vol. A role for the beta-(1,3)galT upstream homolog, jhp0562; 2009. p. 111. [Google Scholar]
- 36.Salaun L, Saunders NJ. Population-associated differences between the phase variable LPS biosynthetic genes of Helicobacter pylori. BMC microbiology. 2006;6:79. doi: 10.1186/1471-2180-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, et al. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999 Jan 14;397(6715):176–80. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 38.Sharma C, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464(7286):250–5. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 39.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997 Aug 7;388(6642):539–47. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 40.Appelmelk BJ, Martin SL, Monteiro MA, Clayton CA, McColm AA, Zheng P, et al. Phase variation in Helicobacter pylori lipopolysaccharide due to changes in the lengths of poly(C) tracts in alpha3-fucosyltransferase genes. Infect Immun. 1999 Oct;67(10):5361–6. doi: 10.1128/iai.67.10.5361-5366.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Appelmelk BJ, Shiberu B, Trinks C, Tapsi N, Zheng PY, Verboom T, et al. Phase variation in Helicobacter pylori lipopolysaccharide. Infect Immun. 1998 Jan;66(1):70–6. doi: 10.1128/iai.66.1.70-76.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Valencia G, Munoz-Perez L, Morales-Espinosa R, Camorlinga-Ponce M, Munoz O, Torres J. Lewis antigen expression by Helicobacter pylori strains colonizing different regions of the stomach of individual patients. J Clin Microbiol. 2008 Aug;46(8):2783–5. doi: 10.1128/JCM.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuipers EJ, Israel DA, Kusters JG, Gerrits MM, Weel J, van Der Ende A, et al. Quasispecies development of Helicobacter pylori observed in paired isolates obtained years apart from the same host. J Infect Dis. 2000 Jan;181(1):273–82. doi: 10.1086/315173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson C, Skoglund A, Moran AP, Annuk H, Engstrand L, Normark S. Lipopolysaccharide diversity evolving in Helicobacter pylori communities through genetic modifications in fucosyltransferases. PLoS ONE. 2008;3(11):e3811. doi: 10.1371/journal.pone.0003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wirth HP, Yang M, Peek RM, Jr, Hook-Nikanne J, Fried M, Blaser MJ. Phenotypic diversity in Lewis expression of Helicobacter pylori isolates from the same host. J Lab Clin Med. 1999 May;133(5):488–500. doi: 10.1016/s0022-2143(99)90026-4. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson C, Skoglund A, Moran AP, Annuk H, Engstrand L, Normark S. An enzymatic ruler modulates Lewis antigen glycosylation of Helicobacter pylori LPS during persistent infection. Proc Natl Acad Sci U S A. 2006 Feb 21;103(8):2863–8. doi: 10.1073/pnas.0511119103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salaun L, Ayraud S, Saunders NJ. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiology. 2005 Mar;151(Pt 3):917–23. doi: 10.1099/mic.0.27379-0. [DOI] [PubMed] [Google Scholar]
- 48.Salaun L, Linz B, Suerbaum S, Saunders NJ. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiology. 2004 Apr;150(Pt 4):817–30. doi: 10.1099/mic.0.26993-0. [DOI] [PubMed] [Google Scholar]
- 49.Sanabria-Valentin E, Colbert MT, Blaser MJ. Role of futC slipped strand mispairing in Helicobacter pylori Lewis(y) phase variation. Microbes Infect. 2007 Nov–Dec;9(14–15):1553–60. doi: 10.1016/j.micinf.2007.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G, Ge Z, Rasko DA, Taylor DE. Lewis antigens in Helicobacter pylori: biosynthesis and phase variation. Mol Microbiol. 2000 Jun;36(6):1187–96. doi: 10.1046/j.1365-2958.2000.01934.x. [DOI] [PubMed] [Google Scholar]
- 51.Sheu B-S, Wu J-J. Type 1 and 2 Lewis antigens of Helicobacter pylori - a potential marker of the human geographical distribution. Journal of medical microbiology. 2008;57(5):543–4. doi: 10.1099/jmm.0.2008/001404-0. [DOI] [PubMed] [Google Scholar]
- 52.Lundin A, Bjrkholm B, Kupershmidt I, Unemo M, Nilsson P, Andersson D, et al. Slow genetic divergence of Helicobacter pylori strains during long-term colonization. Infection and immunity. 2005;73(8):4818–22. doi: 10.1128/IAI.73.8.4818-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghose C, Perez-Perez GI, van Doorn LJ, Dominguez-Bello MG, Blaser MJ. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J Clin Microbiol. 2005 Jun;43(6):2635–41. doi: 10.1128/JCM.43.6.2635-2641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim J, Kim J, Chae S, Cha Y, Park S. High prevalence of multiple strain colonization of Helicobacter pylori in Korean patients: DNA diversity among clinical isolates from the gastric corpus, antrum and duodenum. Korean journal of internal medicine. 2004;19(1):1–9. doi: 10.3904/kjim.2004.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Israel DA, Salama N, Krishna U, Rieger UM, Atherton JC, Falkow S, et al. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(25):14625–30. doi: 10.1073/pnas.251551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang J, Blaser MJ. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat Rev Microbiol. 2006 Nov;4(11):826–36. doi: 10.1038/nrmicro1528. [DOI] [PubMed] [Google Scholar]
- 57.Oleastro M, Cordeiro R, Mnard A, Gomes J. Allelic diversity among Helicobacter pylori outer membrane protein genes homB and homA generated by recombination. Journal of bacteriology. 2010;192(15):3961–8. doi: 10.1128/JB.00395-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pride DT, Blaser MJ. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J Mol Biol. 2002 Feb 22;316(3):629–42. doi: 10.1006/jmbi.2001.5311. [DOI] [PubMed] [Google Scholar]
- 59.Matteo M, Granados G, Prez C, Olmos M, Sanchez C, Catalano M. Helicobacter pylori cag pathogenicity island genotype diversity within the gastric niche of a single host. Journal of medical microbiology. 2007;56(5):664–9. doi: 10.1099/jmm.0.46885-0. [DOI] [PubMed] [Google Scholar]
- 60.Kawai M, Furuta Y, Yahara K, Tsuru T, Oshima K, Handa N, et al. Evolution in an oncogenic bacterial species with extreme genome plasticity: Helicobacter pylori East Asian genomes. BMC microbiology. 2011;11:104. doi: 10.1186/1471-2180-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forsyth MH, Cover TL. Mutational analysis of the vacA promoter provides insight into gene transcription in Helicobacter pylori. Journal of bacteriology. 1999;181(7):2261–6. doi: 10.1128/jb.181.7.2261-2266.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.