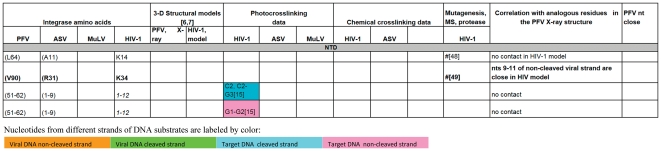
Figure 3. A comparison of all the IN-DNA contact points that have been determined experimentally or modeled for the NTD IN.
The amino acid residues are presented in black, unless a particular residue comes from specified IN monomer in PFV intasome structure as in Refs. [6], [7]. Specific residues shown to interact with DNA that are either in good correlation with the PFV structural results or do not contradict them are bolded. # -this amino acid is in contact with DNA, but the nucleotide is not determined. (G377) - The amino acid residues in parentheses indicate structural analogs to the ones implicated in DNA binding by experimental data. Nucleotides from different strands of DNA substrates are labeled by colors corresponding to the scheme used in Figure 1 and noted above. “G5{15}” - In this example and throughout Figures 3, 4, 5, 6 the nucleotide numbers correspond to the numbering scheme shown in Figure 1 . The numbers in the curly brackets are as in the structure of PFV IN and the model of HIV-1 IN [6], [7]. If listed, the letter designating a nucleotide comes from the original data. All reported contacts are references to original publications with numbers in brackets; our data are marked with asterisks (e.g. A3*).