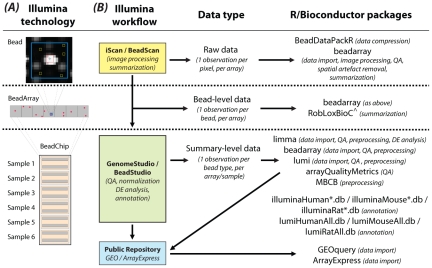
Figure 1. Overview of the technology and workflow.
(A) A zoomed view of a typical bead (top) with the pixels that contribute to the overall (red square) and local background (yellow squares) signals marked. Many replicate beads that contain the same 50-mer oligo are located on each BeadArray (middle) to ensure robust measures of expression can be obtained for each probe in a given sample. Around 48,000 different probe types are assayed in this way per sample. These BeadArrays come from a WG-6 BeadChip (bottom), which is made up of a total of 12 arrays, which are paired to allow transcript abundance to be measured in a total of six samples per BeadChip. (B) Summarizes the various data formats available along with the Illumina workflow associated with the different levels of data. Data can be in raw form, where pixel-level data are available from TIFF images, allowing the complete data processing pipeline, including image analysis, to be carried out in R. The next level, referred to as bead-level, refers to the availability of intensity and location information for individual beads. In this format, a given probe will have a variable number of replicate intensities per sample. Processed data, where replicate intensities have been summarized and outliers removed to give a mean, a measure of variability, and a number of observations per probe in each sample, is the most commonly available format. Summary data are usually obtained directly from Illumina's BeadStudio/GenomeStudio software, but can also be retrieved from public repositories such as GEO or ArrayExpress. The right-hand column of this figure indicates the R/Bioconductor packages that can handle data in these different formats. Probe annotation packages are also listed. List of abbreviations and footnotes used in this figure: QA, quality assessment; DE, differential expression; ∧, package available from CRAN [46]; *, denotes chip-specific part of package name that depends upon platform version (e.g., v1, v2, v3, v4).