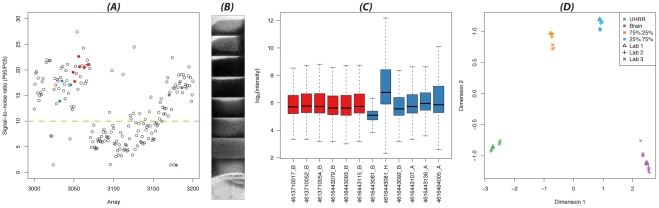
Figure 2. Various diagnostic plots which are useful for quality assessment.
Where scanner metrics information is available, arrays within a particular experiment can be compared to each other, or to a wider set from the same core facility. In (A), a per array signal-to-noise value (95th percentile of signal divided by the 5th percentile) is plotted for 200 consecutive BeadArrays, with the arrays from the experiment in question highlighted in color (blue or red). Low signal-to-noise ratios indicate a poor dynamic range of intensities and can highlight problems with array processing when they occur sequentially over time. At the individual array level, sub-array artefacts can be detected using spatial plots of the intensities across the BeadArray surface (B) and removed using BASH and outlier removal. For a between sample display, boxplots of the intensities from different arrays within an experiment can highlight samples with unusual signal distributions (C). The relationships between different samples can also be assessed using a multi-dimensional scaling (MDS) plot (D), which can highlight true biological differences between samples (in this example, the difference between UHRR and Brain in dimension 1 and the pure versus mixed samples in dimension 2), as well as technical effects due to lab, experiment date, etc., which may also need to be accounted for in the modelling.