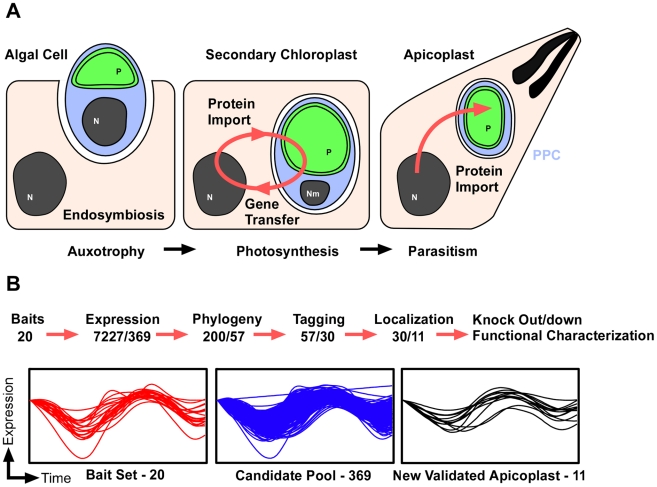
Figure 1. Discovering new apicoplast proteins weighing multiple data sources.
(A) Schematic outline of the acquisition and evolution of the apicoplast. An algal cell (blue) carrying a chloroplast (green) enters an endosymbiotic relationship with a protist host. The establishment of a protein import system allows for massive gene transfer and the development of a stable union. The initial compartments of the endosymbionts are highly conserved over long periods of evolutionary time. PPC, the periplastid compartment is highlighted in blue. (B) Overview of the pipeline used for the identification, prioritization, confirmation and disruption of apicoplast proteins. The graphs show mRNA abundance profiles over two consecutive tachyzoite cell-cycles as described in [36]. Profiles are shown for those proteins used as seeds (left), the resulting candidate pool (middle), and for the newly identified apicoplast proteins (right).